The following models are also available at GitHub: lmrodriguezr/rocker.
§ Index
- AmoA: Ammonia monooxygenase
- AmoA_B_v2 (bacterial, v2.0)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Comments: Explicitly excludes non-bacterial variants
Last update: Apr-18-2017 - AmoA_A_v2 (archaeal, v2.0)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Comments: Explicitly excludes non-archaeal variants
Last update: Apr-18-2017 - AmoA_B (bacterial)
- 6 models: 80bp, 100bp, 125bp, 150bp, 200bp, 250bp
Prepared by: Luis (Coto) Orellana
Comments: Explicitly excludes non-bacterial variants
Last update: Nov-06-2015 - AmoA_A (archaeal)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Comments: Explicitly excludes non-archaeal variants
Last update: Nov-13-2015
- BLA: Beta-lactamase
- BlaA
- 2 models: 150bp, 250bp
Prepared by: Siyu Zhang
Comments: Class A beta-lactamase [with--search diamond]
Last update: Apr-19-2020 - MBL
- 2 models: 150bp, 250bp
Prepared by: Brittany Suttner
Comments: Metalo-beta-lactamase (class B) [with--search diamond]
Last update: Feb-18-2020 - MBL_S1_S2
- 2 models: 150bp, 250bp
Prepared by: Brittany Suttner
Comments: Metalo-beta-lactamase (class B) subclasses S1 and S2 [with--search diamond]
Last update: Feb-18-2020 - MBL_S3
- 2 models: 150bp, 250bp
Prepared by: Brittany Suttner
Comments: Metalo-beta-lactamase (class B) subclass S3 [with--search diamond]
Last update: Feb-18-2020 - BlaC
- 2 models: 150bp, 250bp
Prepared by: Siyu Zhang
Comments: Class C beta-lactamase
Last update: Feb-24-2020 - OXA
- 2 models: 150bp, 250bp
Prepared by: Brittany Suttner
Comments: Beta-lactamase OXA family (blaOXA, class D) [with--search diamond]
Last update: Feb-18-2020 - TEM
- 2 models: 150bp, 250bp
Prepared by: Siyu Zhang
Comments: Beta-lactamase TEM family (blaTEM) [with--search diamond]
Last update: Feb-24-2020
- Hao: Hydroxylamine oxidoreductase
- Hao
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
- KSalpha: Ketosynthase alpha subunit
- KSalpha
- 1 model: 150bp
Prepared by: Brittany Suttner
Comments: Polyketide antibiotic biosynthesis
Last update: Nov-20-2018
- NarG: Nitrate reductase
- NarG
- 6 models: 80bp, 100bp, 150bp, 200bp, 250bp, 500bp
Prepared by: Despina Tsementzi & Luis M Rodriguez
Comments: [with--search diamond]
Last update: Jun-11-2015
- NarG_NxrA: Nitrate reductase and nitrite oxidoreductase alpha subunit
- NarG_NxrA
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
- NifH: Nitrogenase reductase subunit
- NifH
- 3 models: 100bp, 150bp, 200bp
Prepared by: Luis M Rodriguez-R and John Christian Gaby
Last update: Aug-04-2018
- NirK: Nitrite reductase
- NirK
- 6 models: 80bp, 100bp, 150bp, 200bp, 250bp, 500bp
Prepared by: Luis (Coto) Orellana
Last update: Jul-14-2015 - NirK_cladeThau (clade Thaumarchaeota)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017 - NirK_cladeIII (clade III)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017 - NirK_cladeI-II (clades I and II)
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
- NorB: Nitric oxide reductase
- NorB
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
- NosZ: Nitrous oxide reductase
- NrfA: Nitrite reductase, formate-dependent
- NrfA
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
- OxyT: N,N-dimethyltransferase
- OxyT
- 1 model: 150bp
Prepared by: Brittany Suttner
Comments: Tetracycline biosynthesis
Last update: Apr-13-2018
- RpoB: Ribosomal RNA polymerase B subunit
- RpoB
- 6 models: 80bp, 100bp, 150bp, 200bp, 250bp, 500bp
Prepared by: Luis M Rodriguez-R
Comments: [with--search diamond]
Last update: Jun-09-2015
- TetM: Tetracycline resistance
- TetM
- 2 models: 100bp, 150bp
Prepared by: Brittany Suttner
Last update: Apr-10-2018
- UreC: Urease subunit alpha
- UreC
- 1 model: 125bp
Prepared by: Luis (Coto) Orellana
Last update: Apr-18-2017
§ Models
FNR: False Negative Rate (1-Sensitivity), FPR: False Positive Rate or fall-out (1-Specificity), Acc: Accuracy.
| Model Name | Length | Statistics | ROCker Plot | ROCker model | Training Set | Comments |
|---|---|---|---|---|---|---|
| AmoA: Ammonia monooxygenase | ||||||
| AmoA_B_v2 | 125bp | FNR: 7.4% FPR: 0.36% Acc: 98.1% | 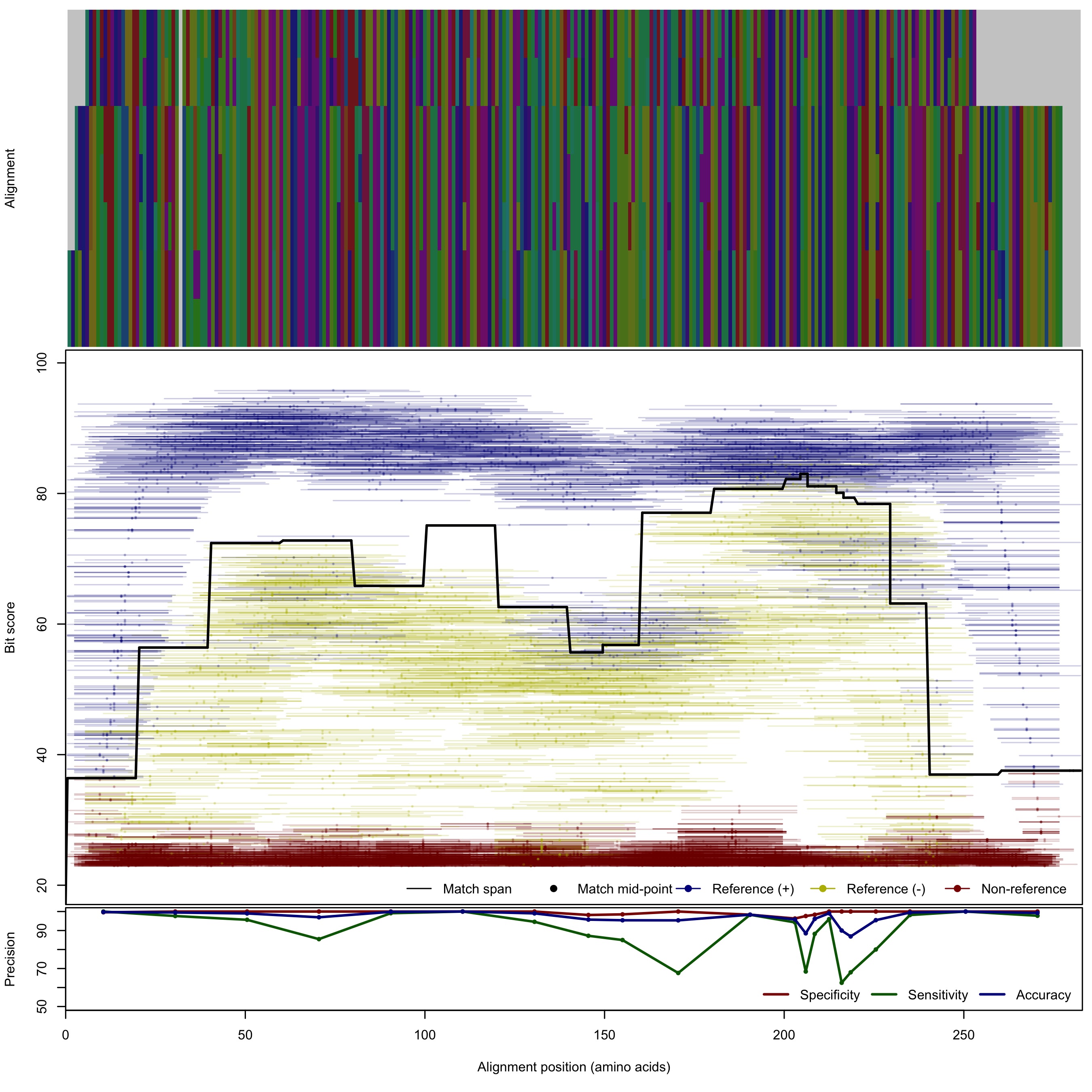 | Download Search online | Positive: 8 proteins Negative: 14 proteins Reference sequences | Explicitly excludes non-bacterial variants Last update: Apr-18-2017 |
| AmoA_A_v2 | 125bp | - | 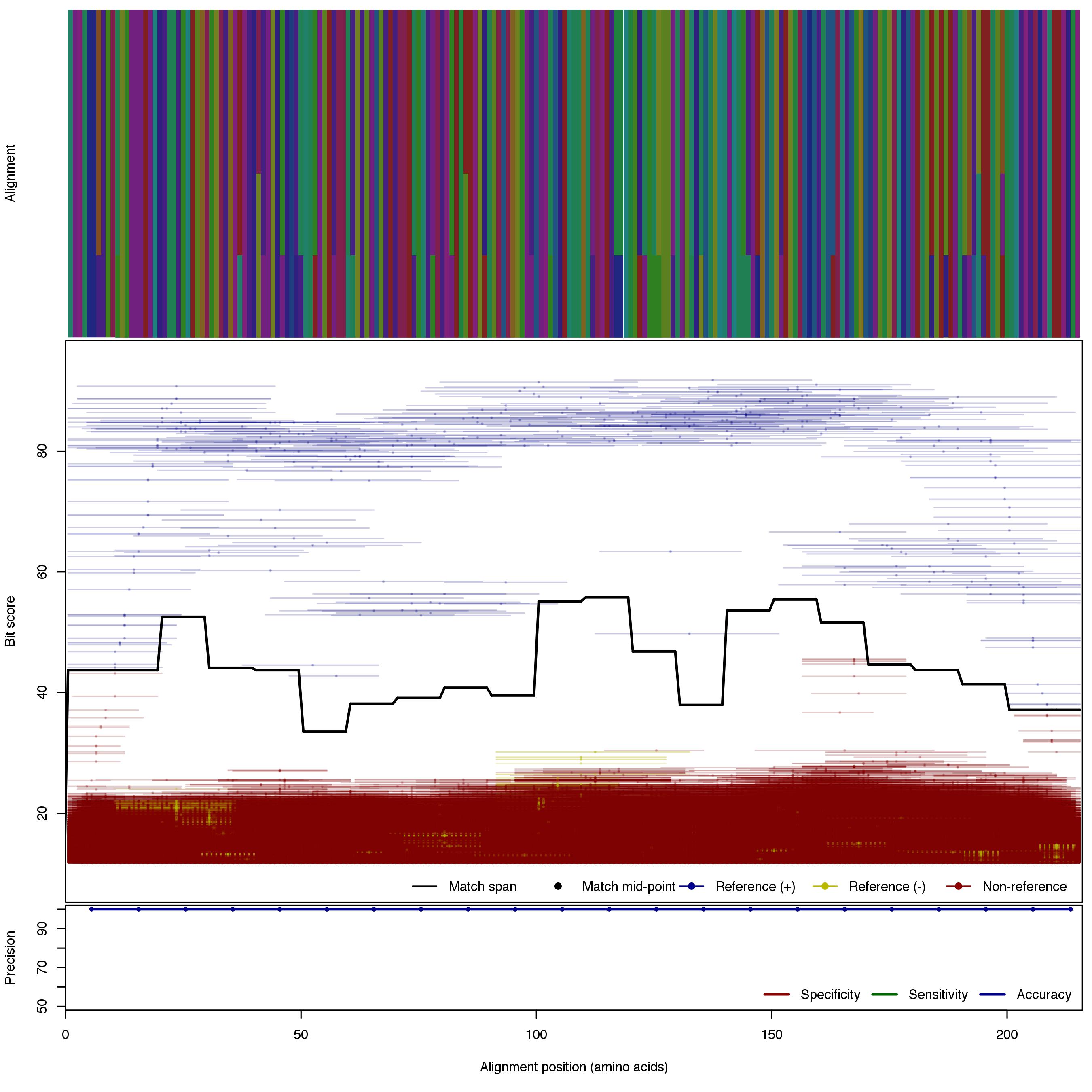 | Download Search online | Positive: 5 proteins Negative: 16 proteins Reference sequences | Explicitly excludes non-archaeal variants Last update: Apr-18-2017 |
| AmoA_B | 80bp | - | 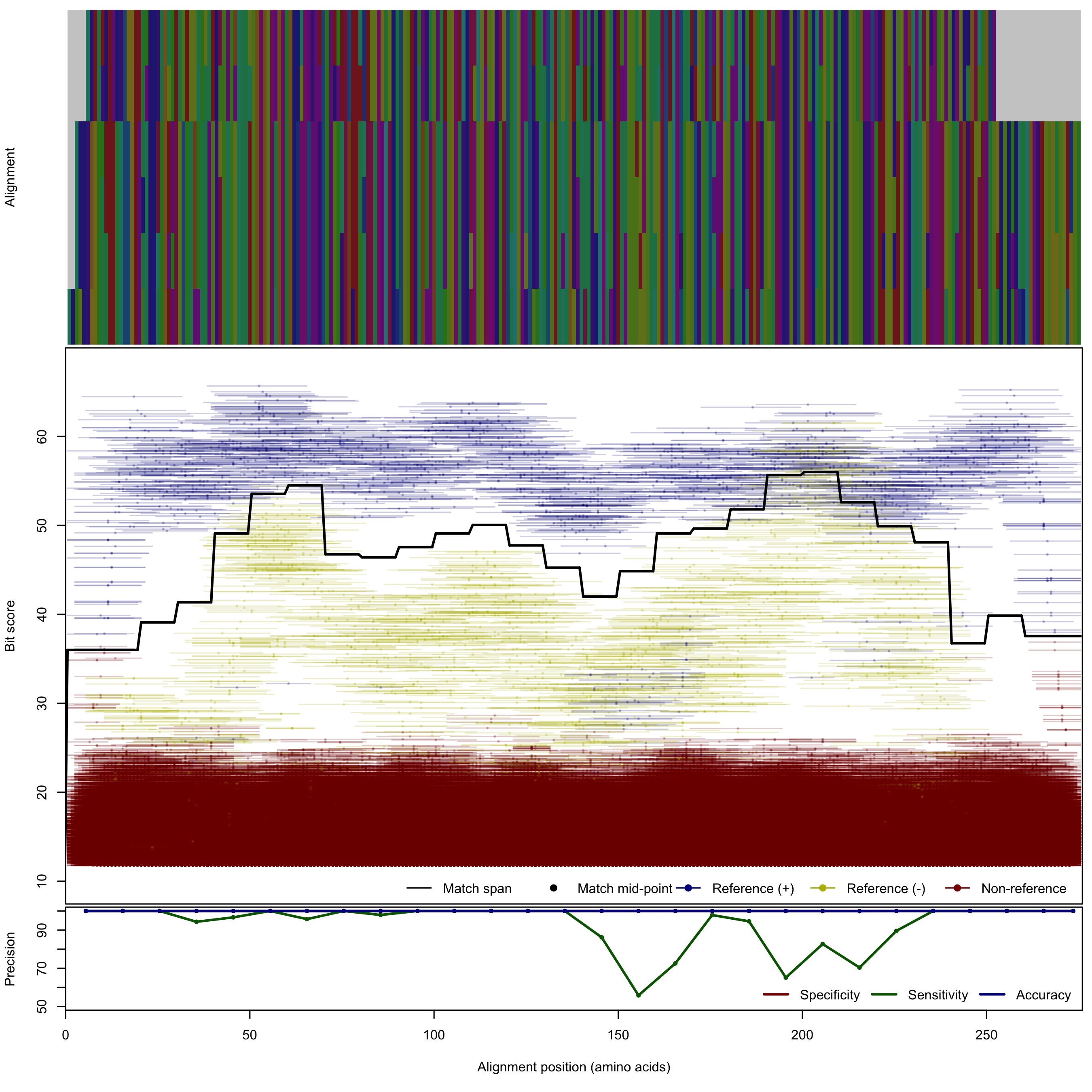 | Download Search online | Positive: 7 proteins Negative: 14 proteins Reference sequences | Explicitly excludes non-bacterial variants Last update: Nov-06-2015 |
| 100bp | - | 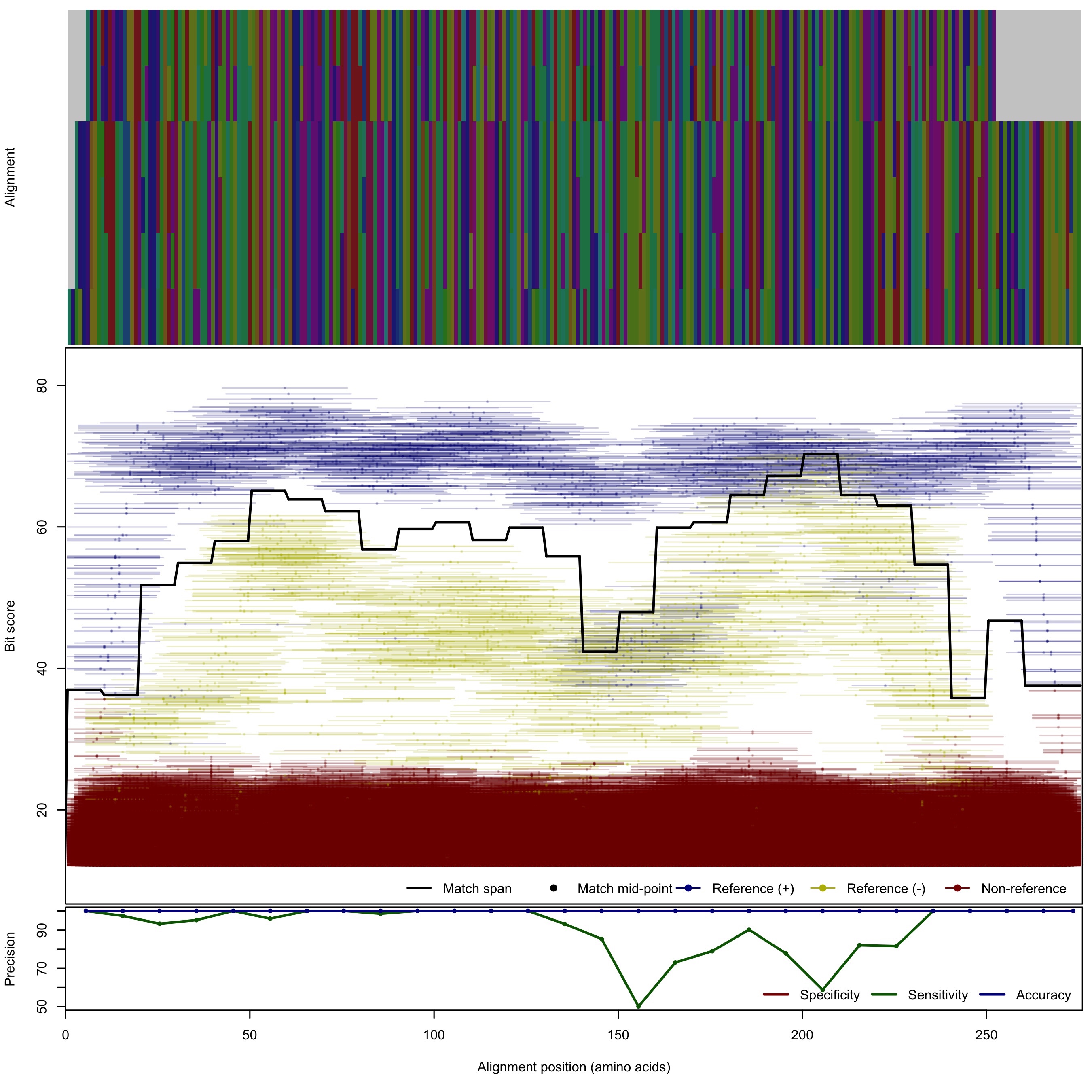 | Download Search online | |||
| 125bp | - | 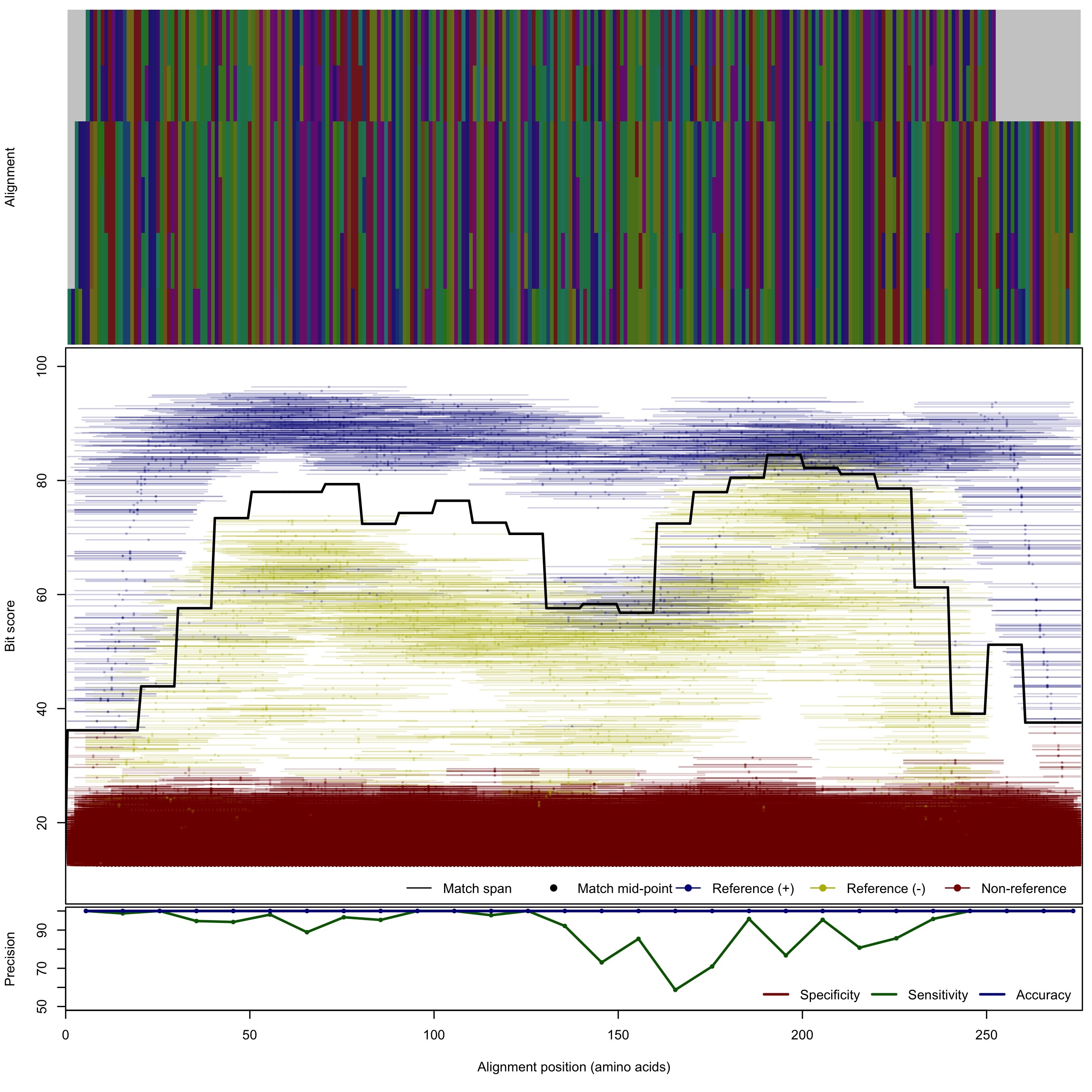 | Download Search online | |||
| 150bp | - | 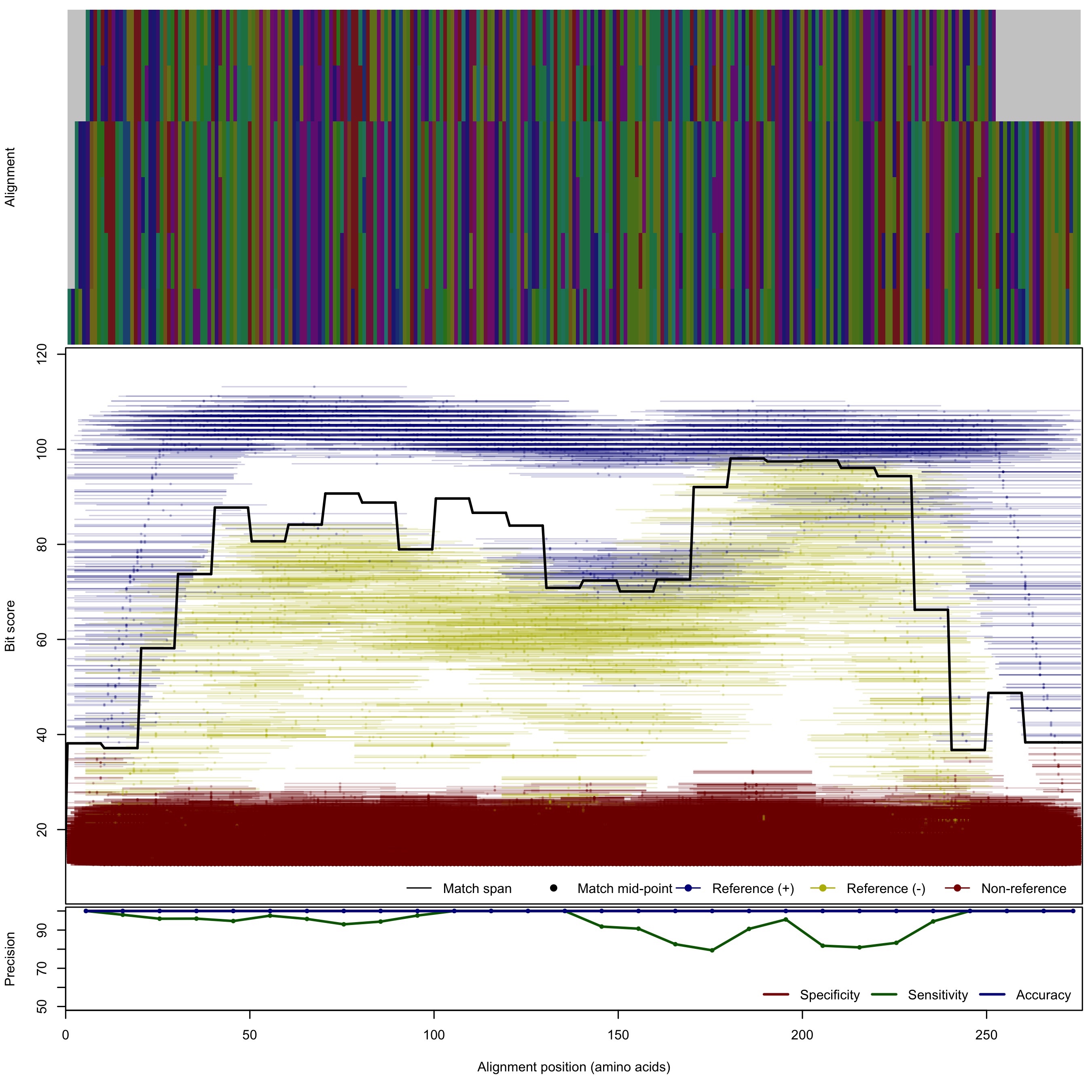 | Download Search online | |||
| 200bp | - | 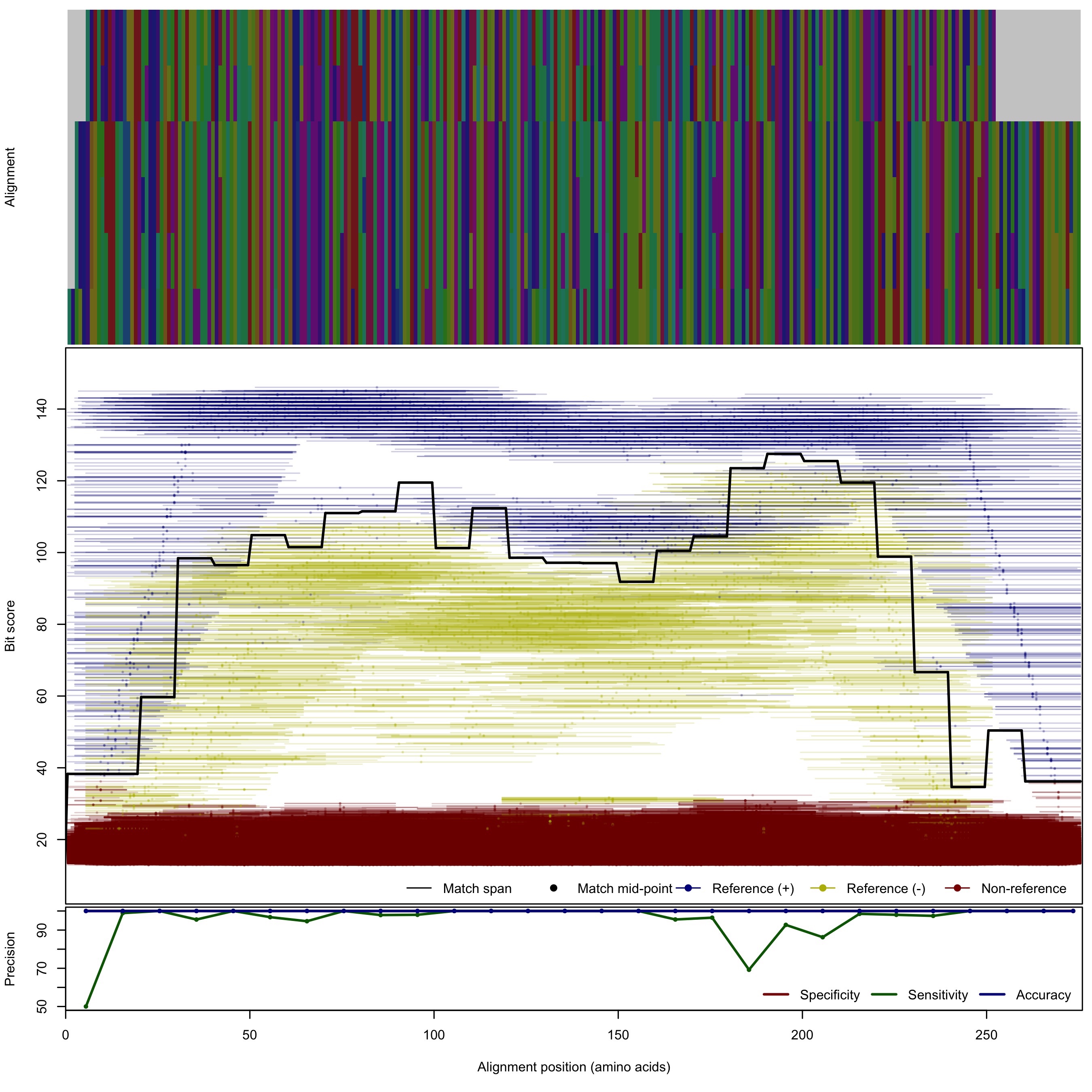 | Download Search online | |||
| 250bp | - | 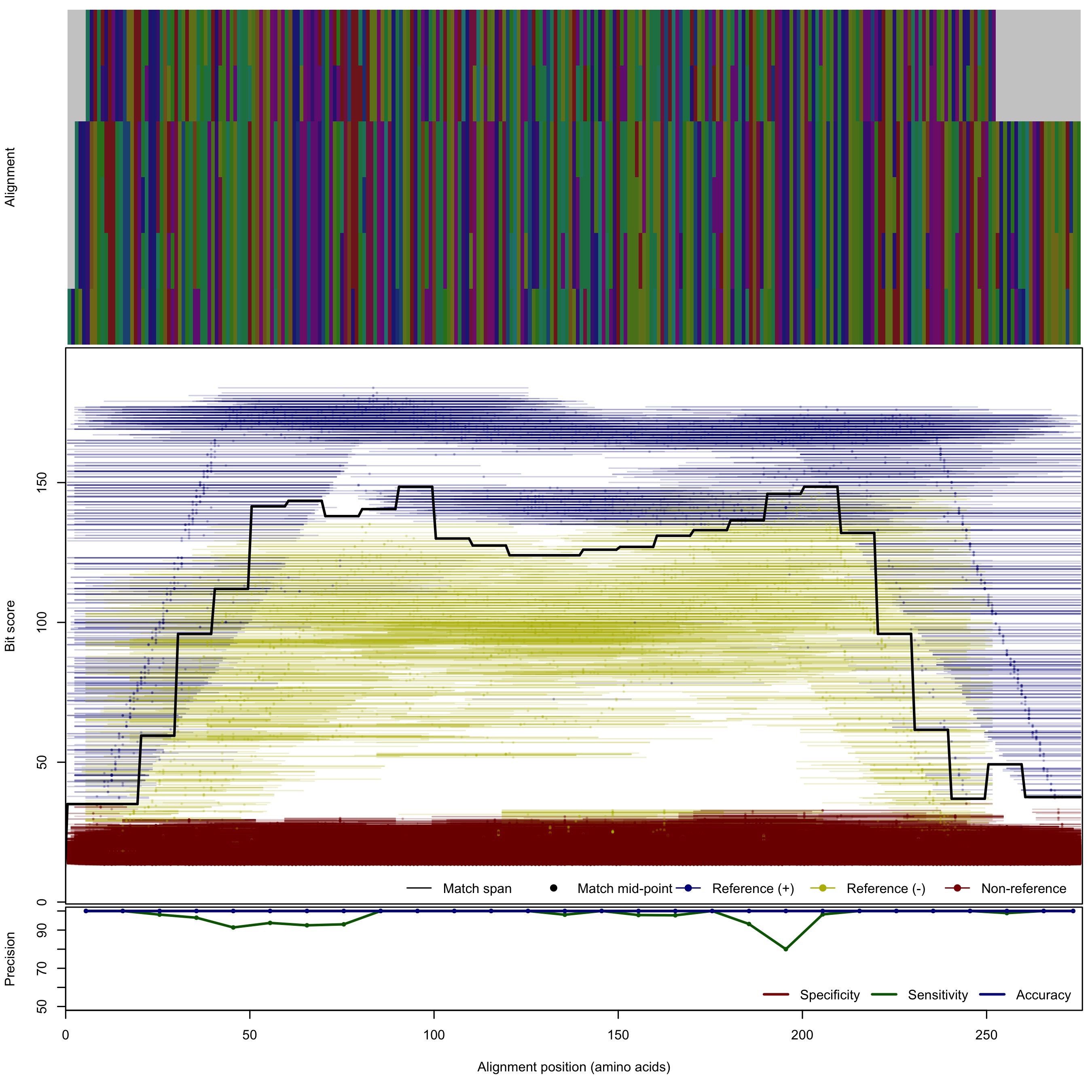 | Download Search online | |||
| AmoA_A | 125bp | - | 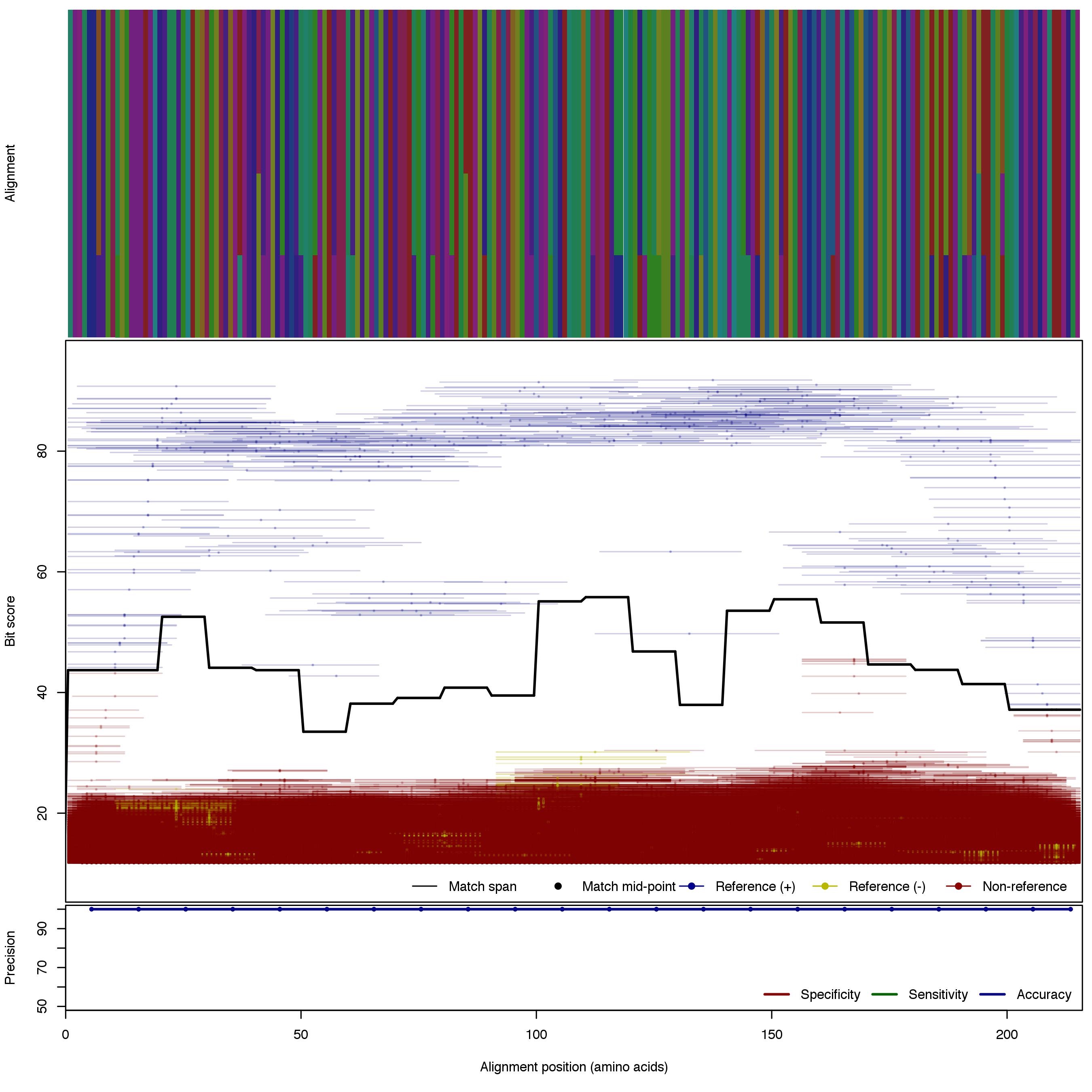 | Download Search online | Positive: 5 proteins Negative: 16 proteins Reference sequences | Explicitly excludes non-archaeal variants Last update: Nov-13-2015 |
| BLA: Beta-lactamase | ||||||
| BlaA | 150bp | FNR: 1.9422% FPR: 0.0055% Acc: 99.982% | 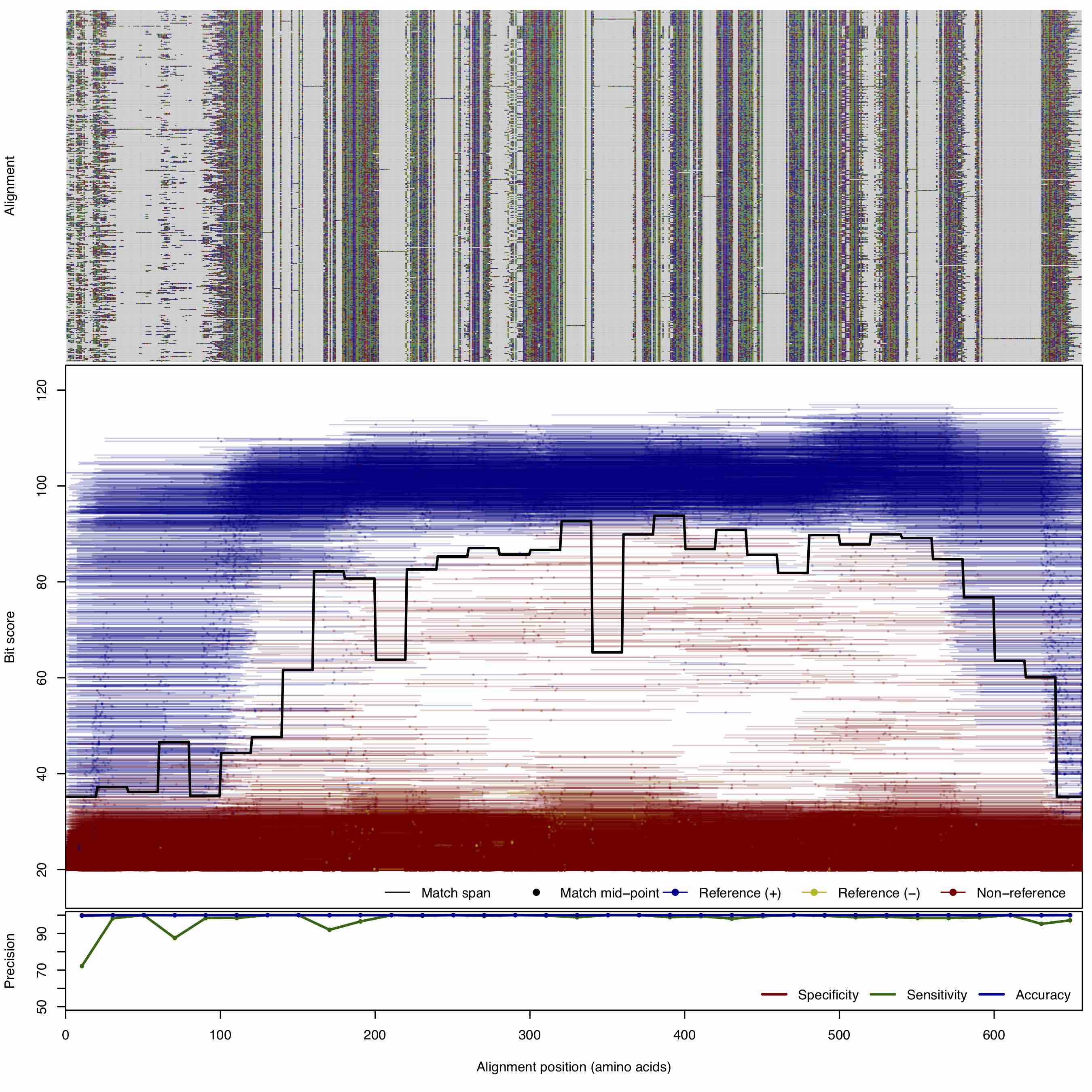 | Download Search online | Positive: 353 proteins Negative: 84 proteins Reference sequences (Trimmed) | Class A beta-lactamase [with --search diamond]Last update: Apr-19-2020 |
| 250bp | FNR: 2.5372% FPR: 0.0042% Acc: 99.987% | 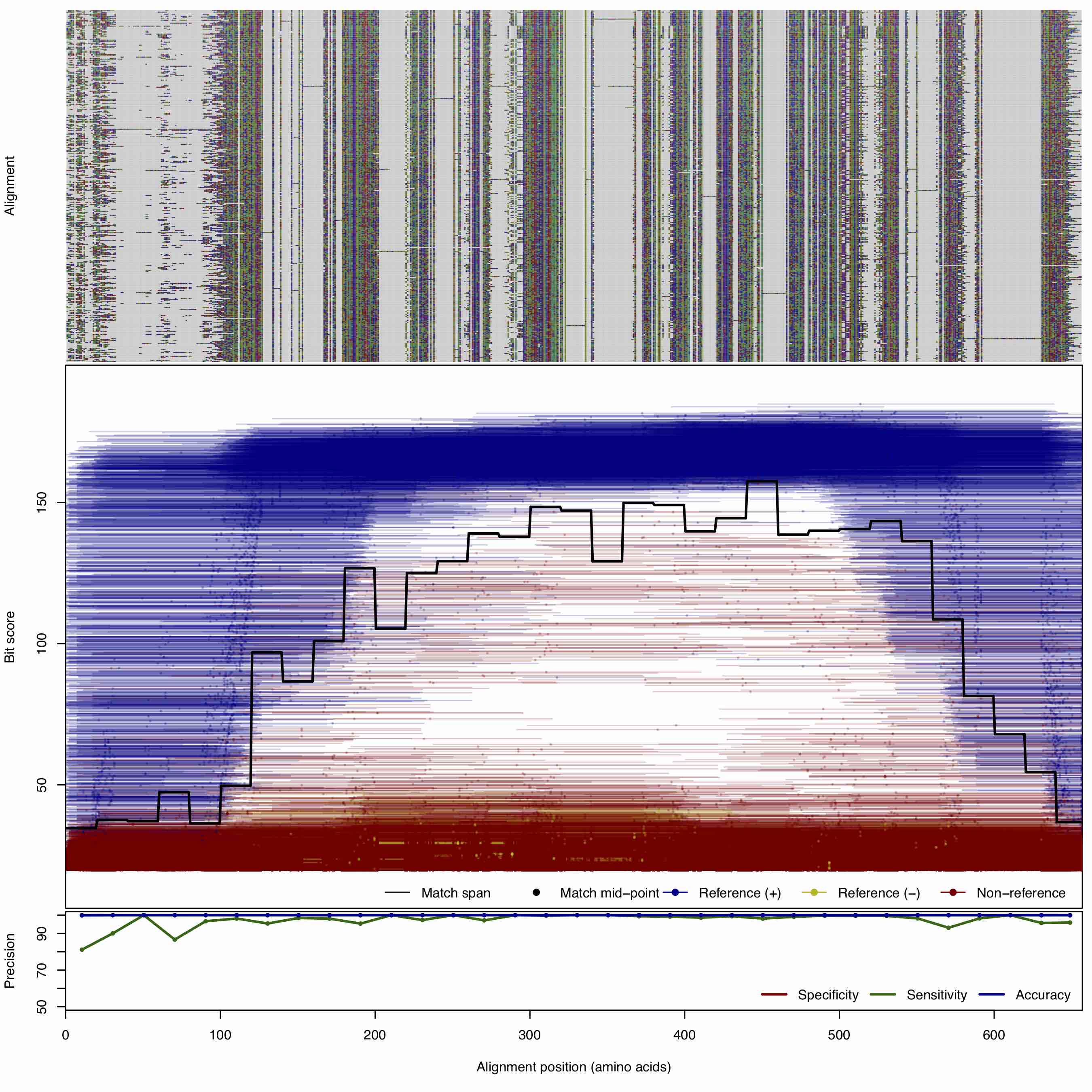 | Download Search online | |||
| MBL | 150bp | FNR: 2.71% FPR: 0.045% Acc: 99.8% | 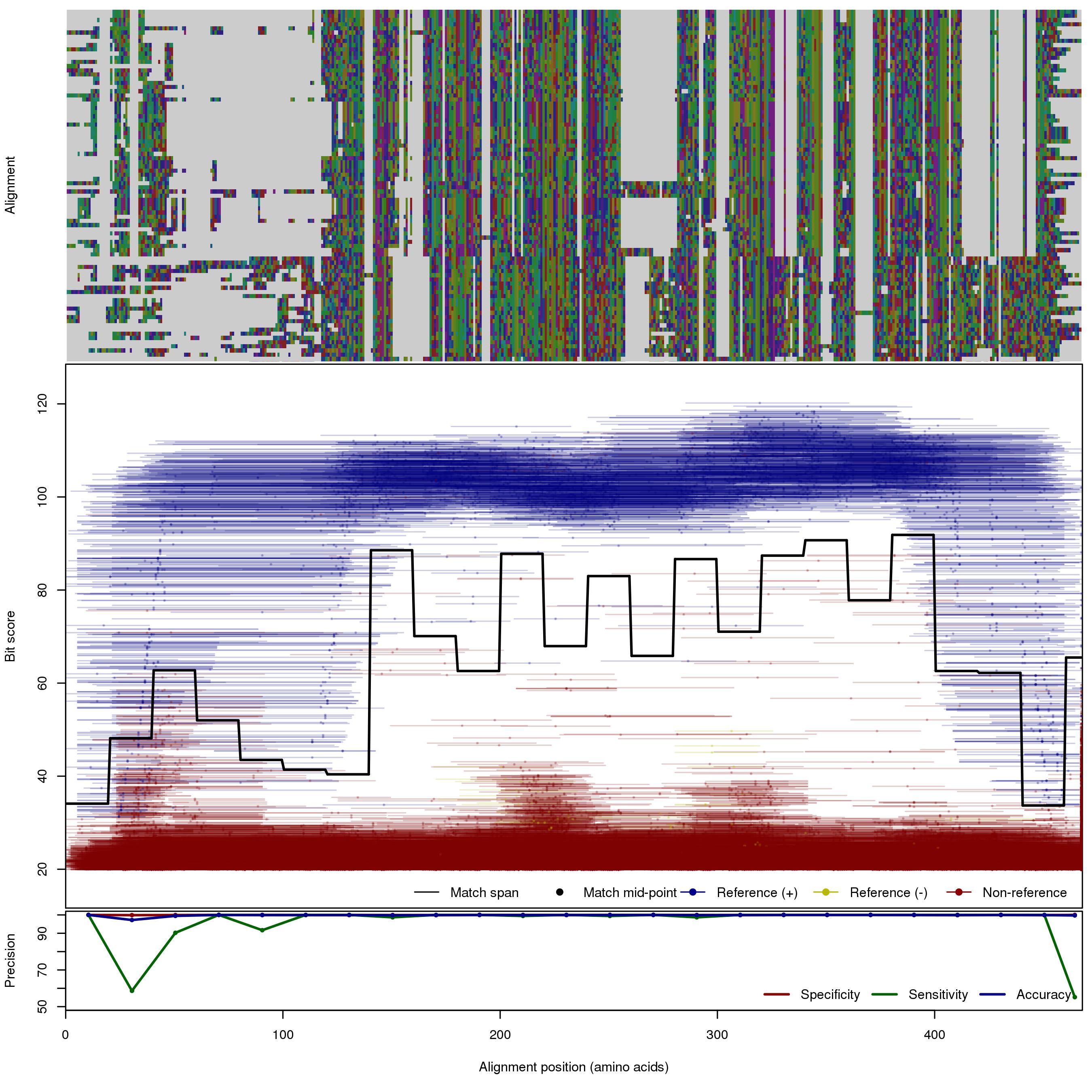 | Download Search online | Positive: 86 proteins Negative: 5 proteins Reference sequences (Trimmed) | Metalo-beta-lactamase (class B) [with --search diamond]Last update: Feb-18-2020 |
| 250bp | FNR: 2.51% FPR: 0.045% Acc: 99.9% | 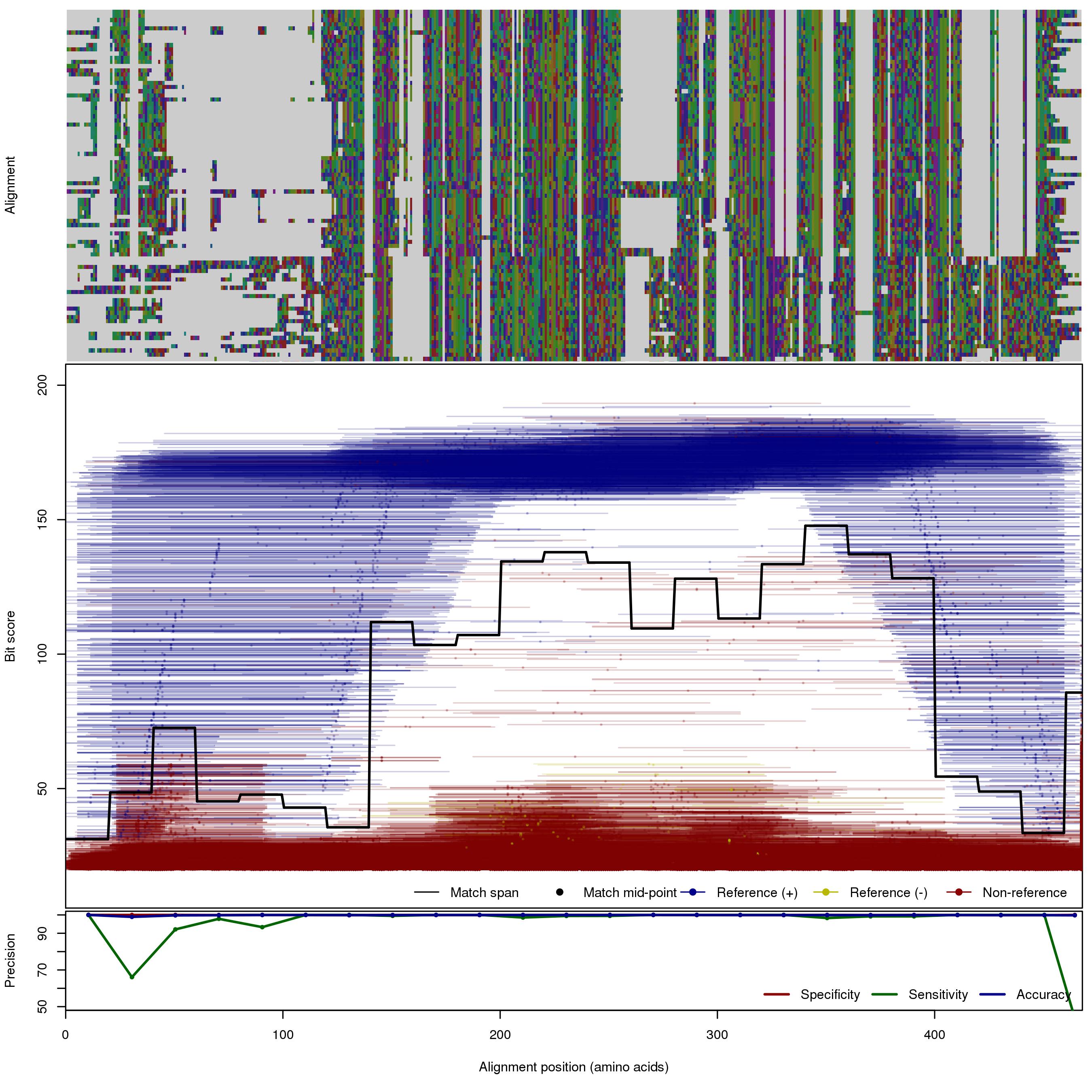 | Download Search online | |||
| MBL_S1_S2 | 150bp | FNR: 1.01% FPR: 0.17% Acc: 99.7% | 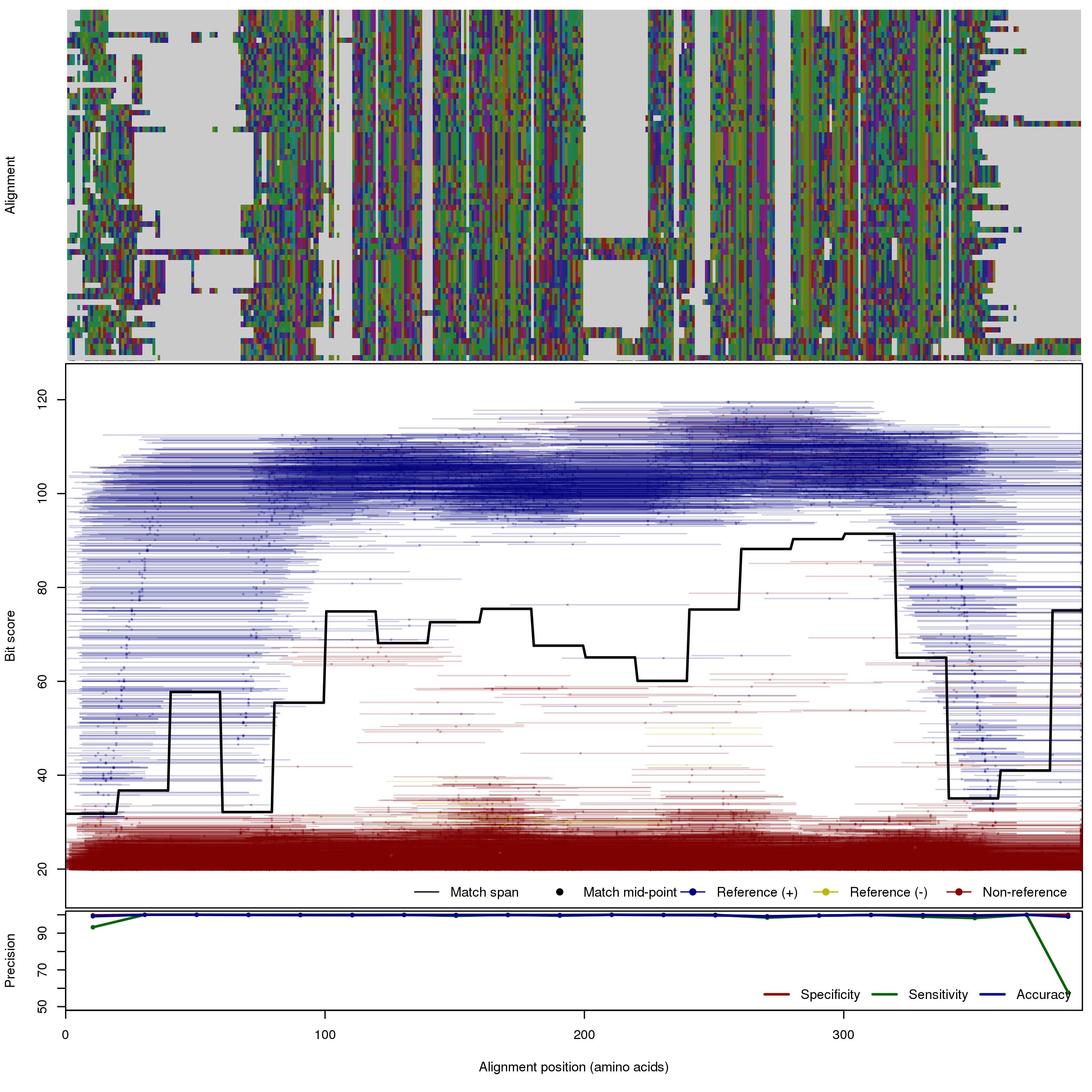 | Download Search online | Positive: 65 proteins Negative: 5 proteins Reference sequences (Trimmed) | Metalo-beta-lactamase (class B) subclasses S1 and S2 [with --search diamond]Last update: Feb-18-2020 |
| 250bp | FNR: 0.631% FPR: 0.0897% Acc: 99.9% | 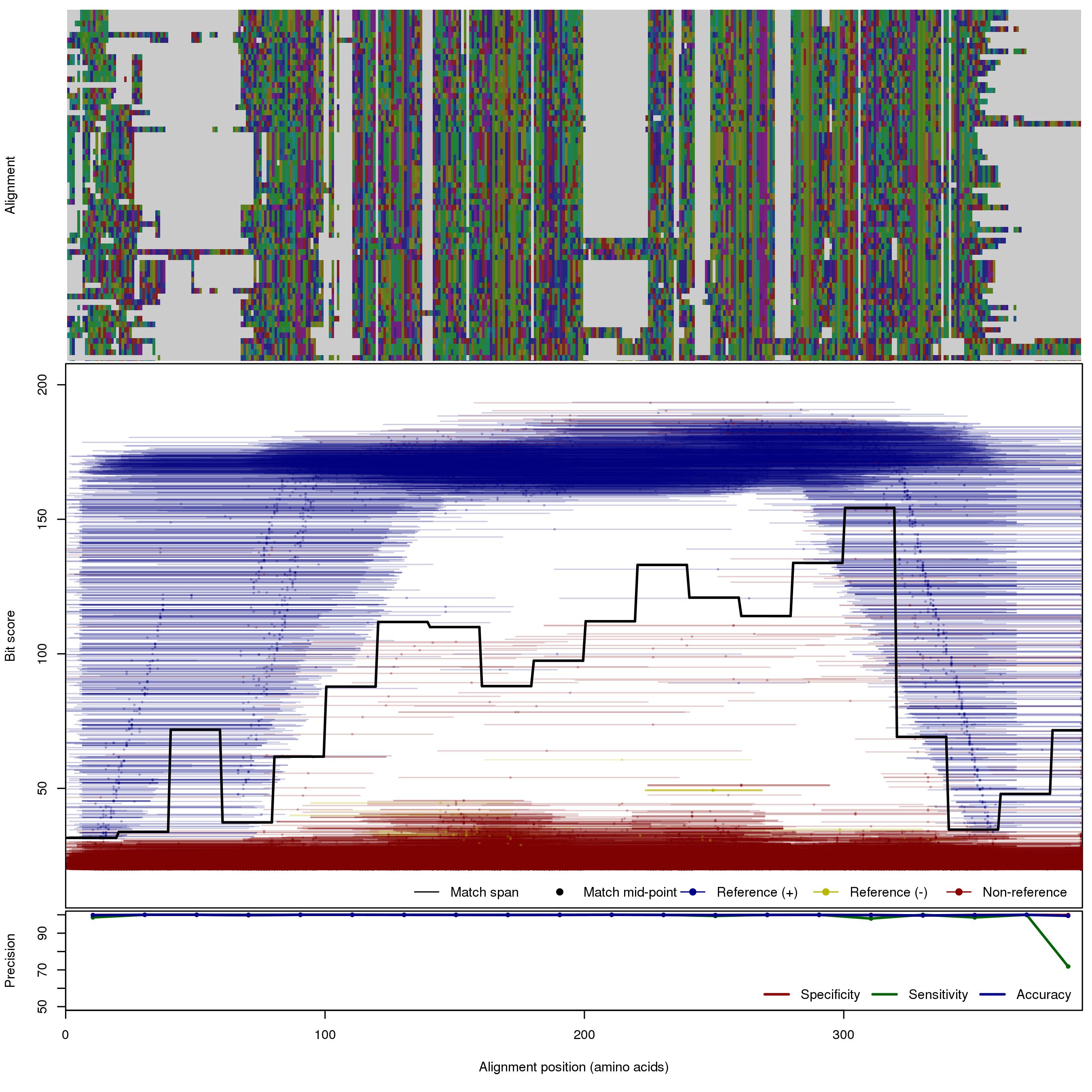 | Download Search online | |||
| MBL_S3 | 150bp | FNR: 5.72% FPR: 0.237% Acc: 99.6% | 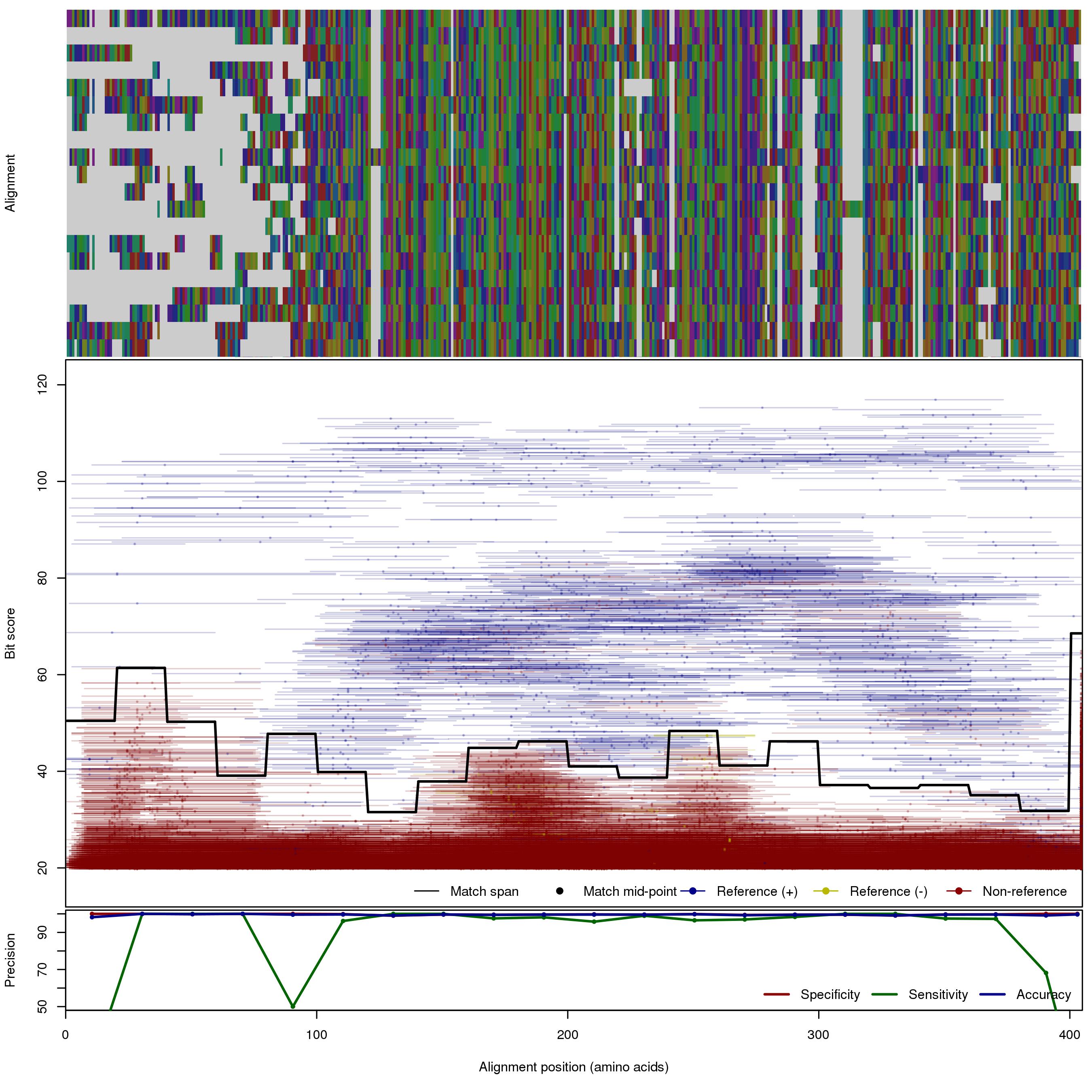 | Download Search online | Positive: 71 proteins Negative: 5 proteins Reference sequences (Trimmed) | Metalo-beta-lactamase (class B) subclass S3 [with --search diamond]Last update: Feb-18-2020 |
| 250bp | FNR: 3.44% FPR: 2.87% Acc: 97.1% | 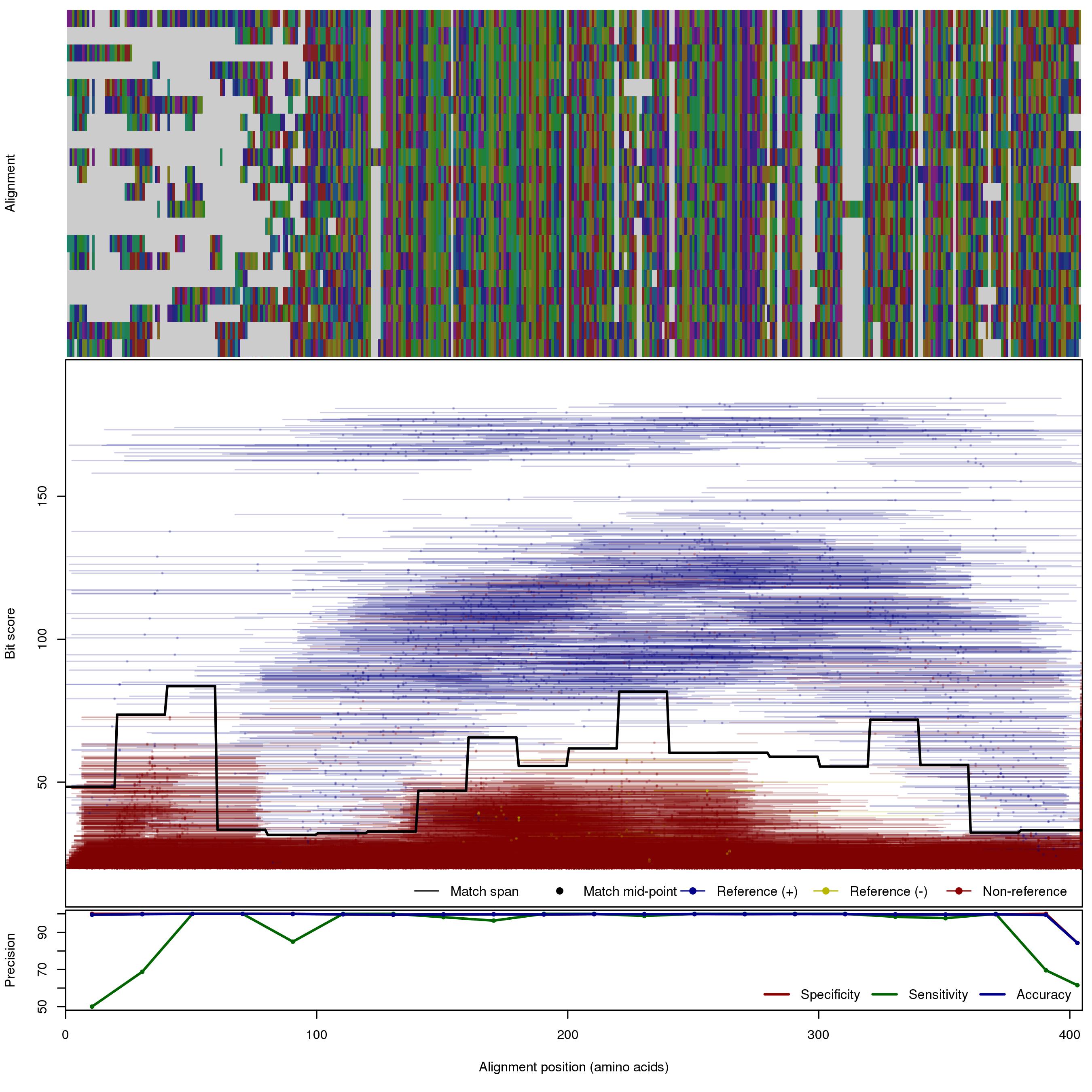 | Download Search online | |||
| BlaC | 150bp | FNR: 1.864% FPR: 9.0E-5% Acc: 99.999% | 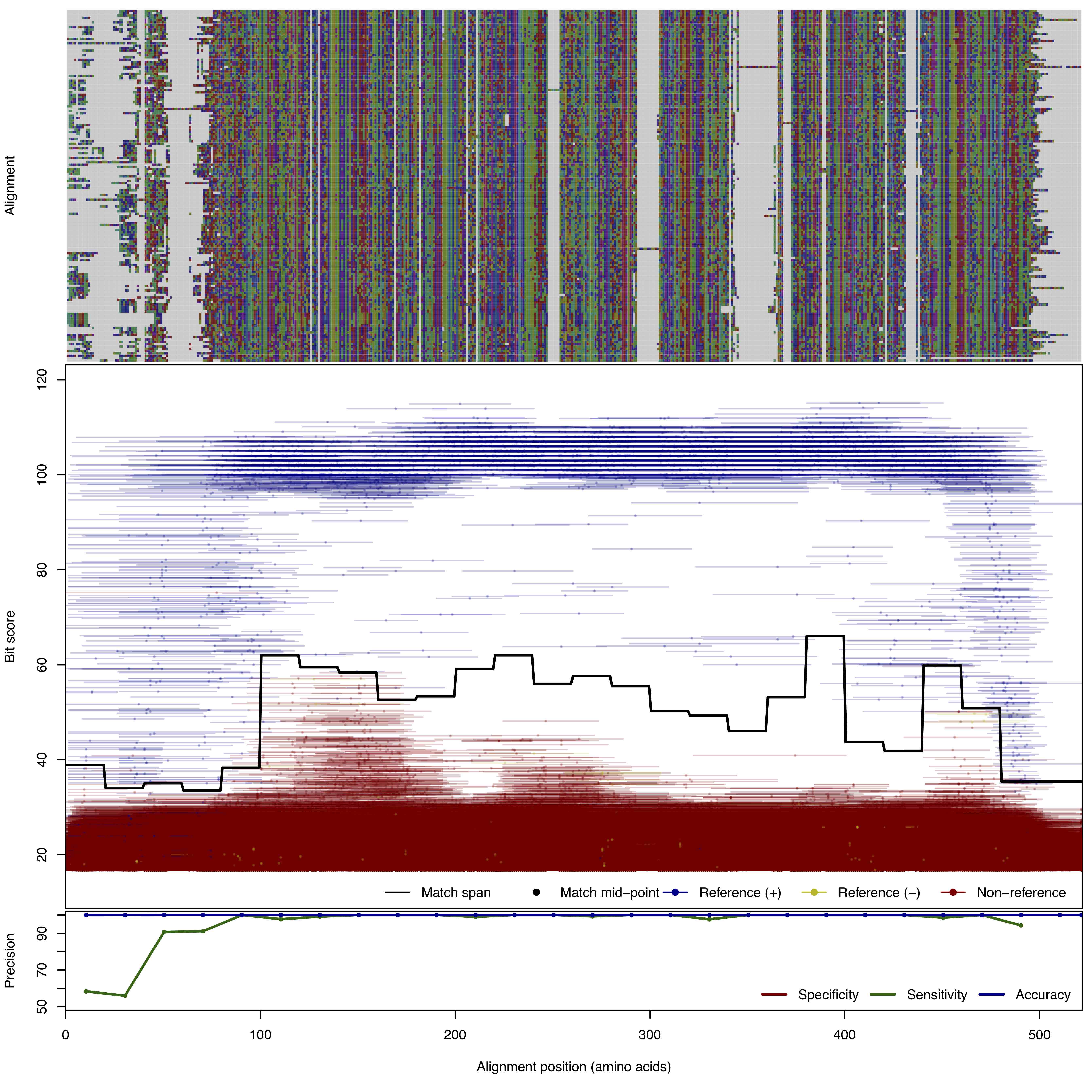 | Download Search online | Positive: 152 proteins Negative: 44 proteins Reference sequences (Trimmed) | Class C beta-lactamase Last update: Feb-24-2020 |
| 250bp | FNR: 2.166% FPR: 0.00022% Acc: 99.999% | 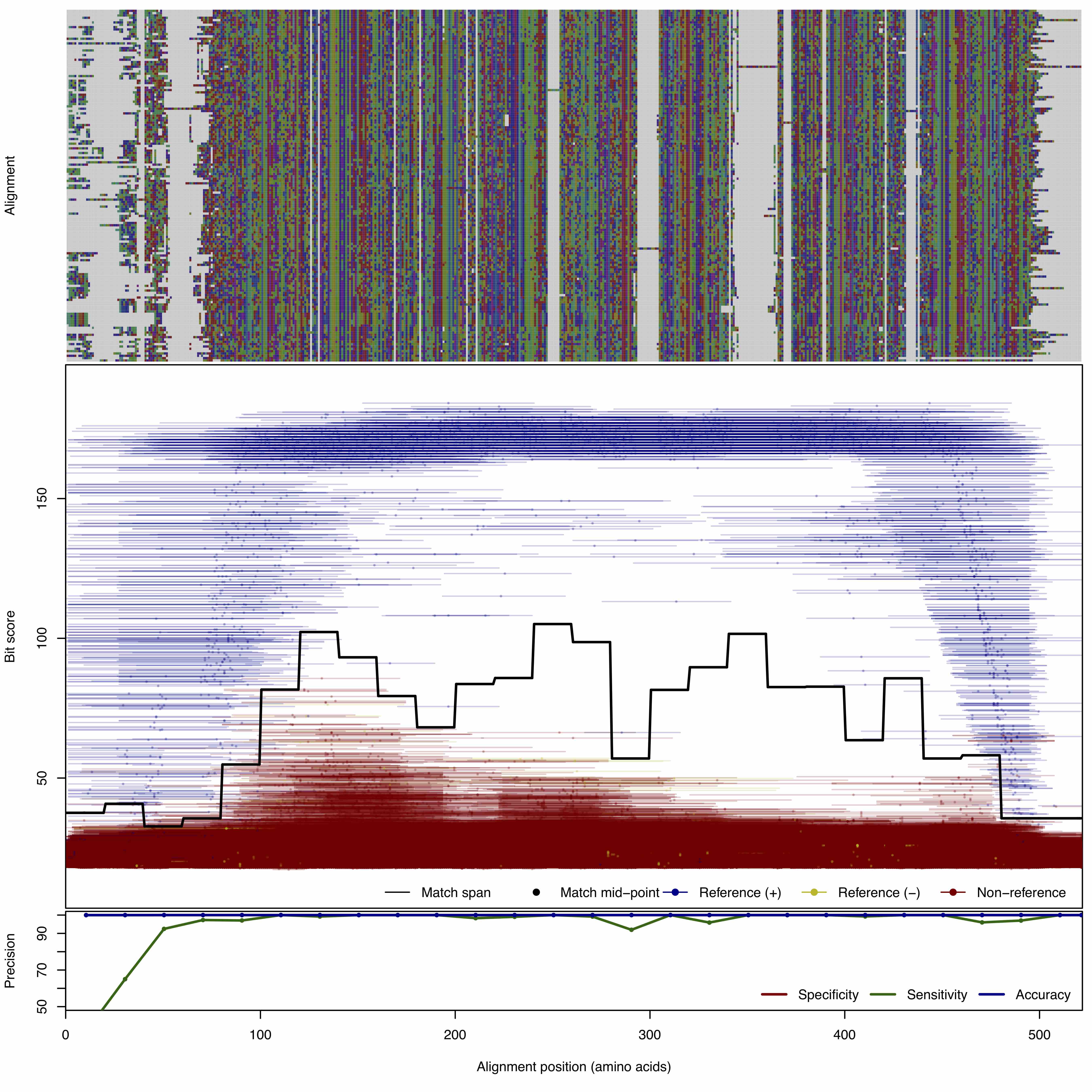 | Download Search online | |||
| OXA | 150bp | FNR: 0.878% FPR: 0.174% Acc: 99.8% | 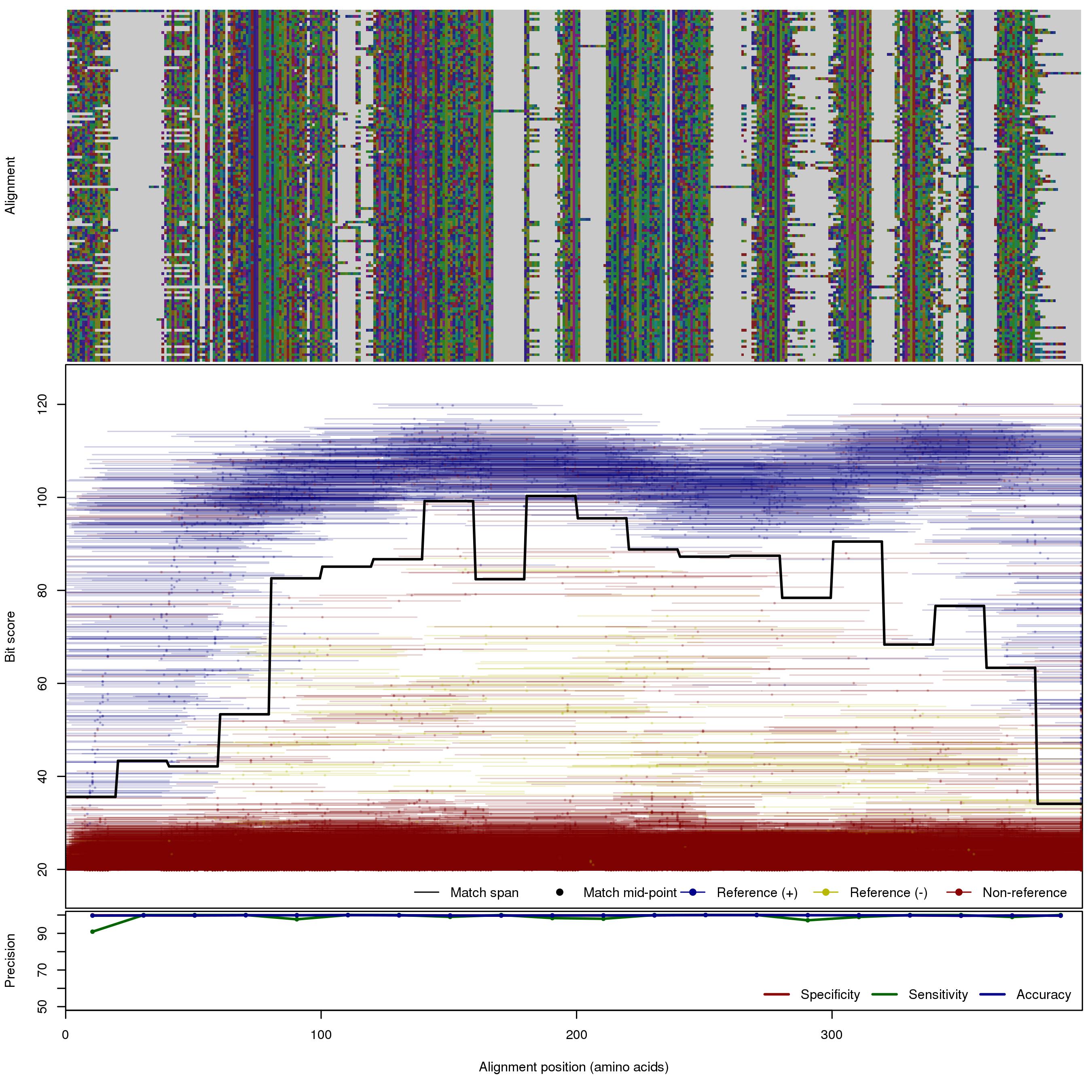 | Download Search online | Positive: 136 proteins Negative: 30 proteins Reference sequences (Trimmed) | Beta-lactamase OXA family (blaOXA, class D) [with --search diamond]Last update: Feb-18-2020 |
| 250bp | FNR: 2.58% FPR: 0.0898% Acc: 99.9% | 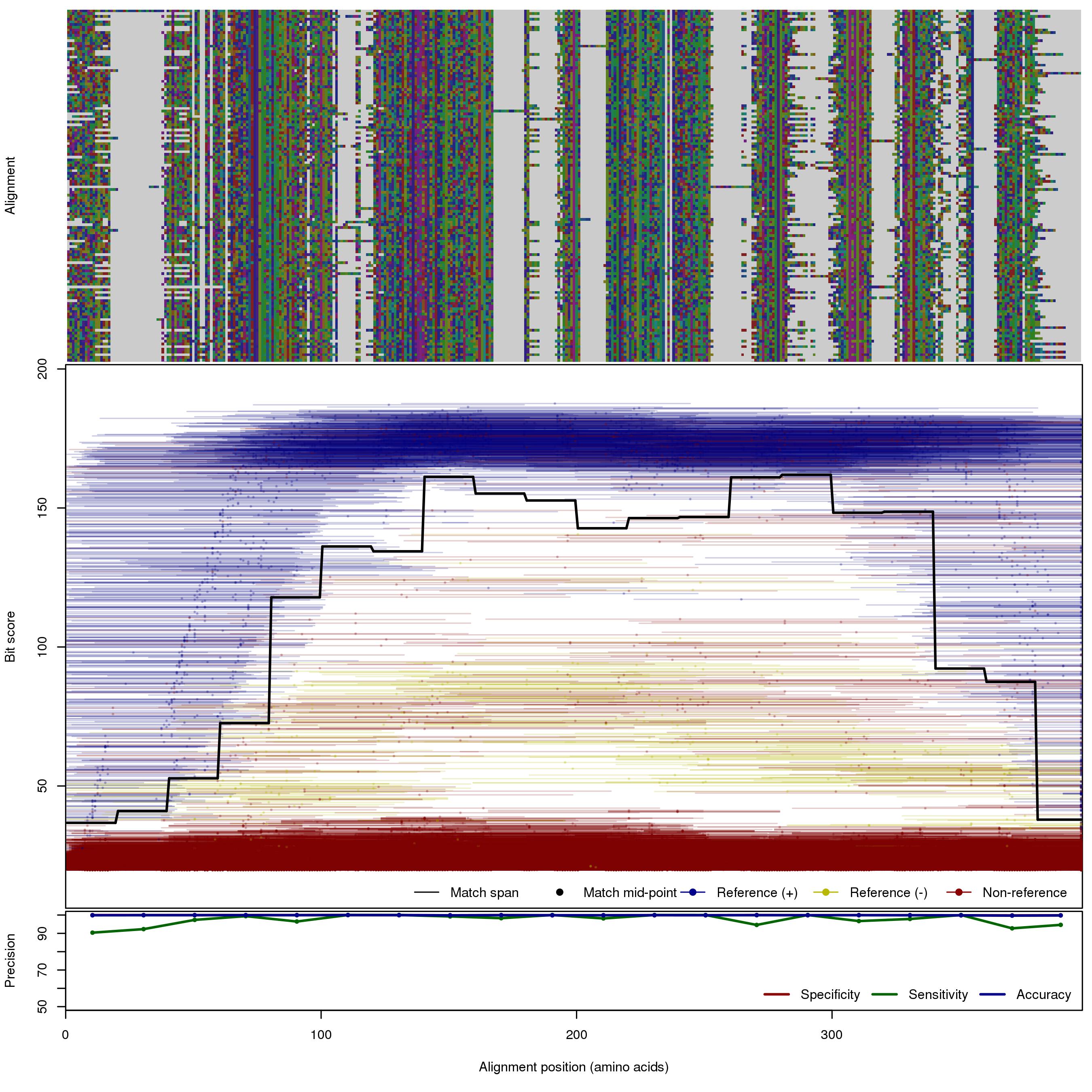 | Download Search online | |||
| TEM | 150bp | FNR: 1.186% FPR: 0.818% Acc: 98.98% | 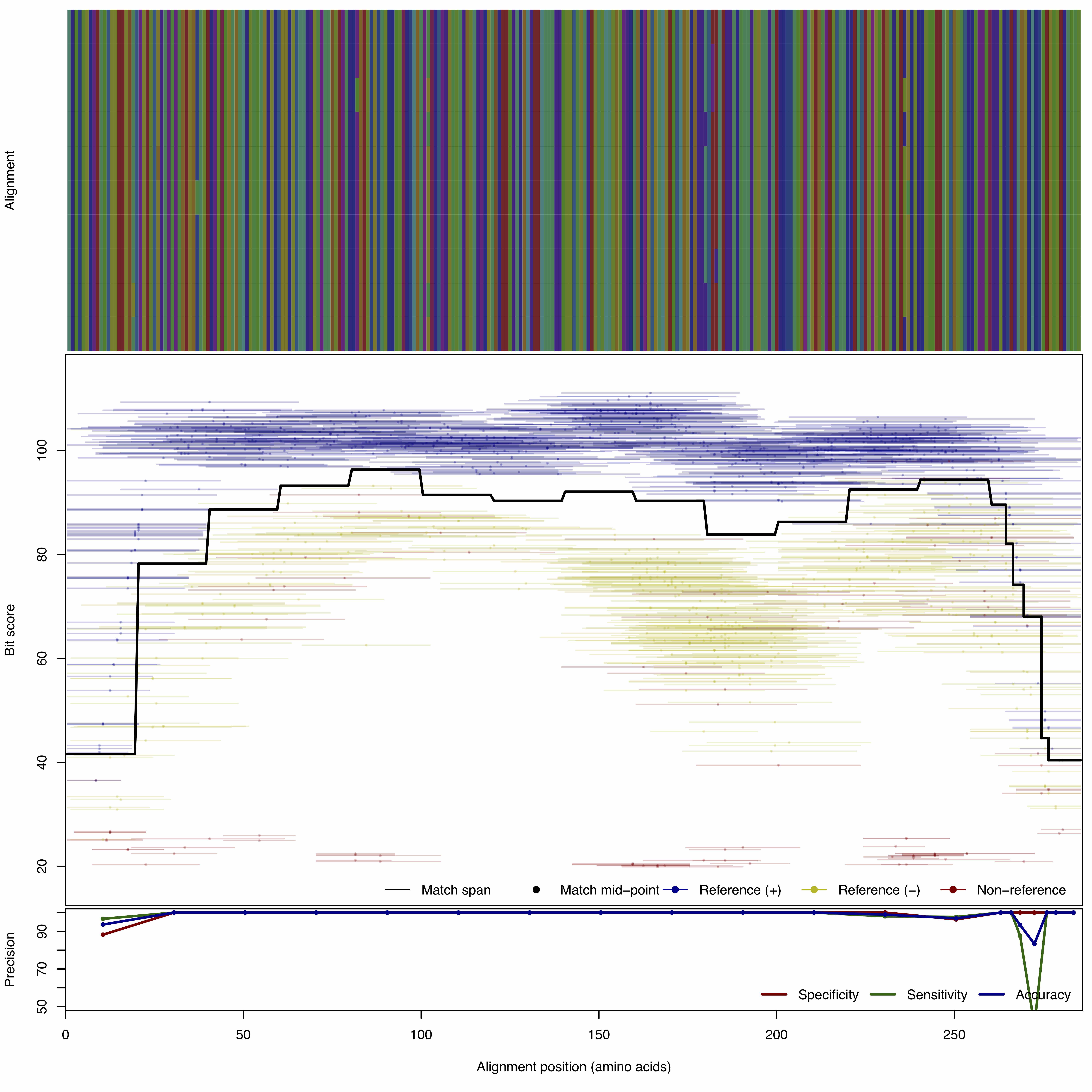 | Download Search online | Positive: 11 proteins Negative: 33 proteins Reference sequences (Trimmed) | Beta-lactamase TEM family (blaTEM) [with --search diamond]Last update: Feb-24-2020 |
| 250bp | FNR: 3.524% FPR: 0.722% Acc: 97.89% | 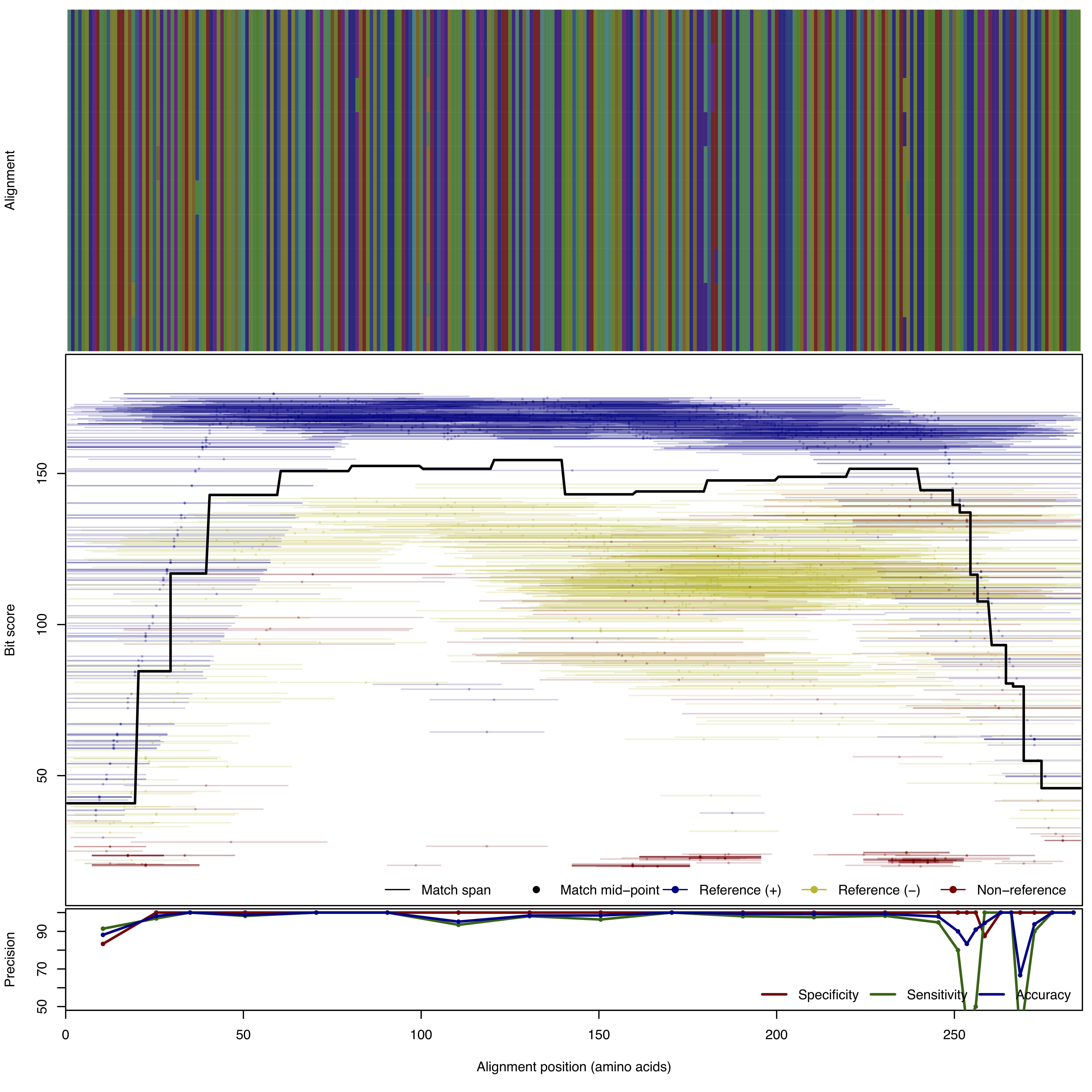 | Download Search online | |||
| Hao: Hydroxylamine oxidoreductase | ||||||
| Hao | 125bp | FNR: 1.28% FPR: 0.11% Acc: 99.14% | 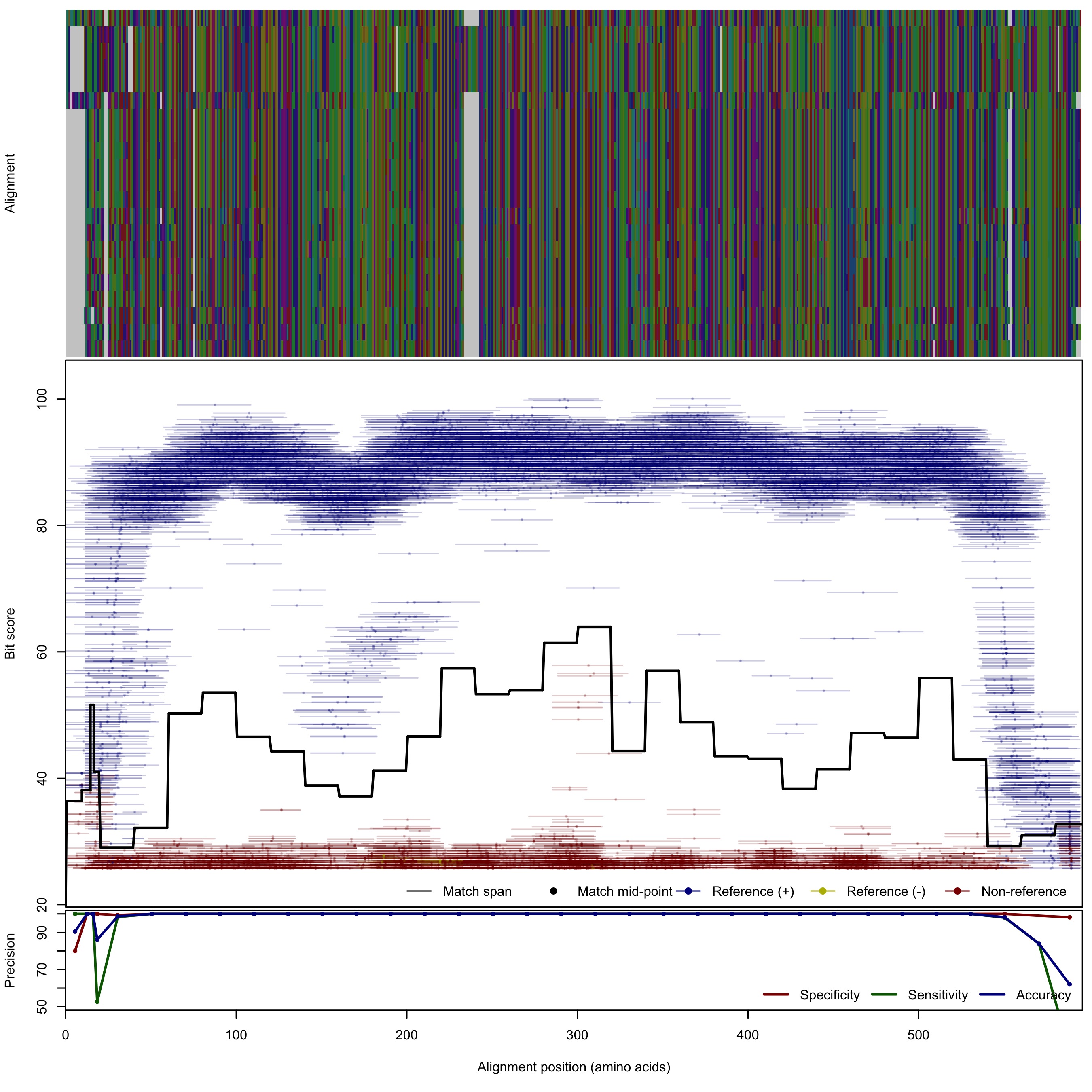 | Download Search online | Positive: 23 proteins Negative: 9 proteins Reference sequences | Last update: Apr-18-2017 |
| KSalpha: Ketosynthase alpha subunit | ||||||
| KSalpha | 150bp | - | 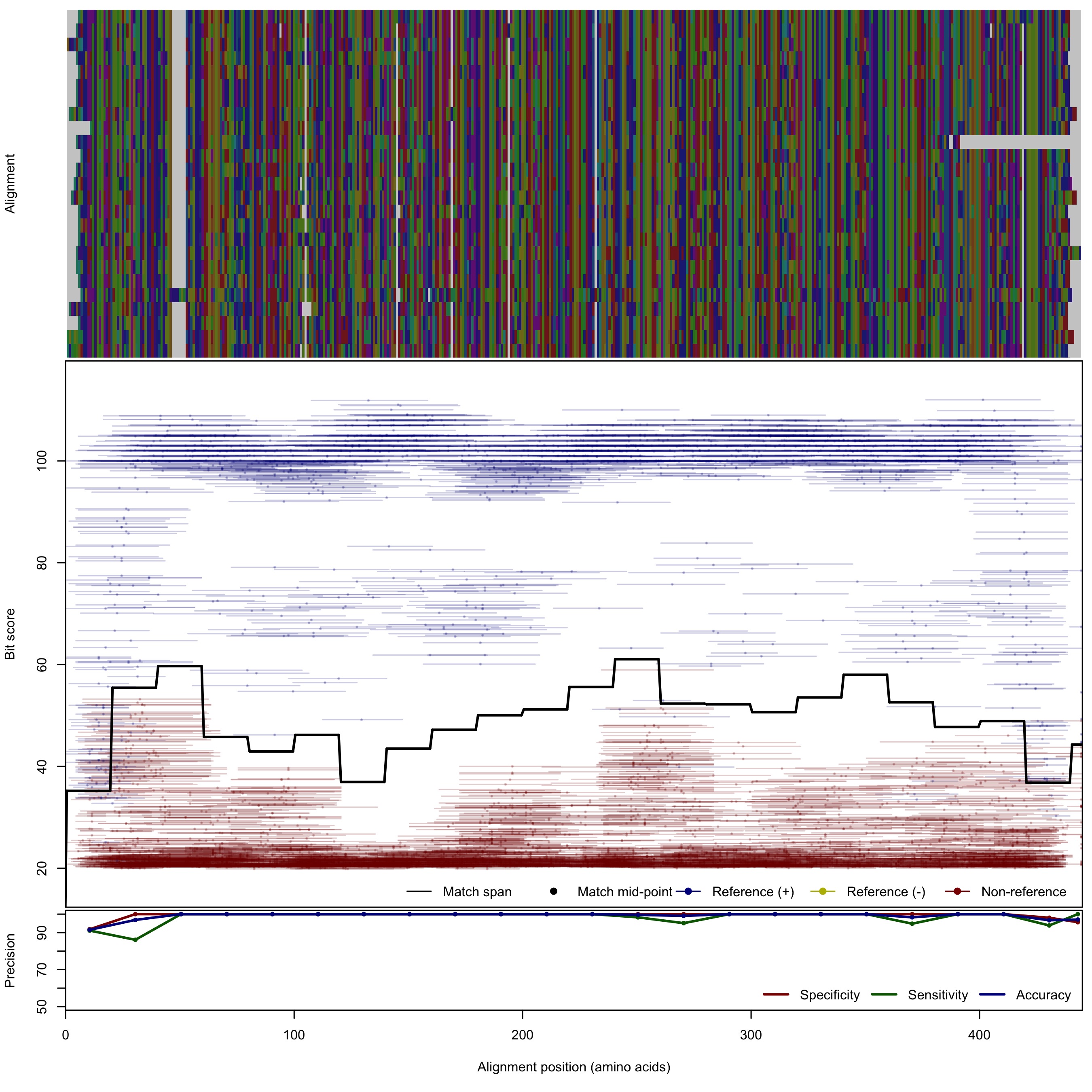 | Download Search online | Positive: 26 proteins Reference sequences | Polyketide antibiotic biosynthesis Last update: Nov-20-2018 |
| NarG: Nitrate reductase | ||||||
| NarG | 80bp | FNR: 0.1% FPR: 11.41% Acc: 97.81% | 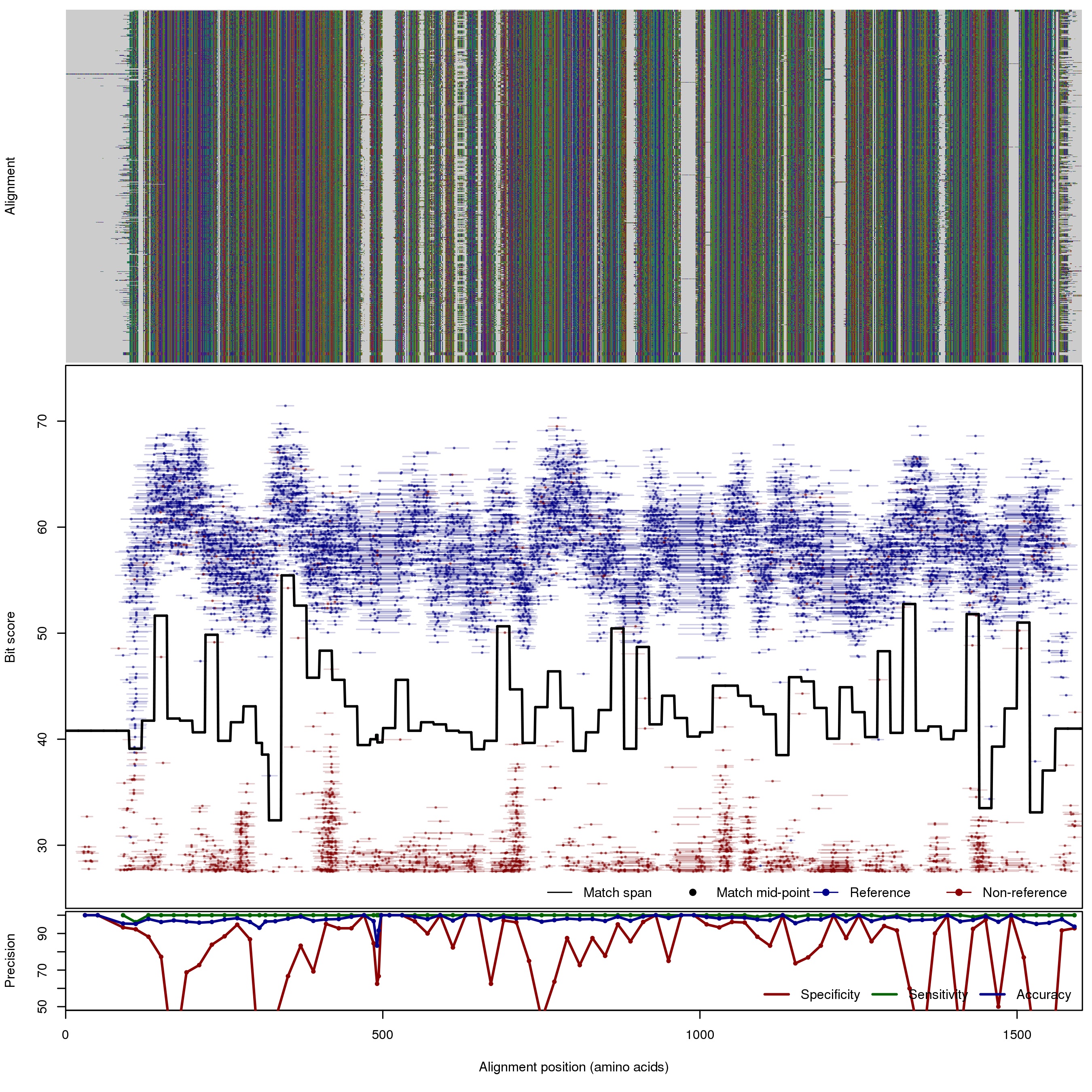 | Download Search online | Positive: 679 proteins Negative: 31 proteins Reference sequences | [with --search diamond]Last update: Jun-11-2015 |
| 100bp | FNR: 0.98% FPR: 5.3% Acc: 97.58% | 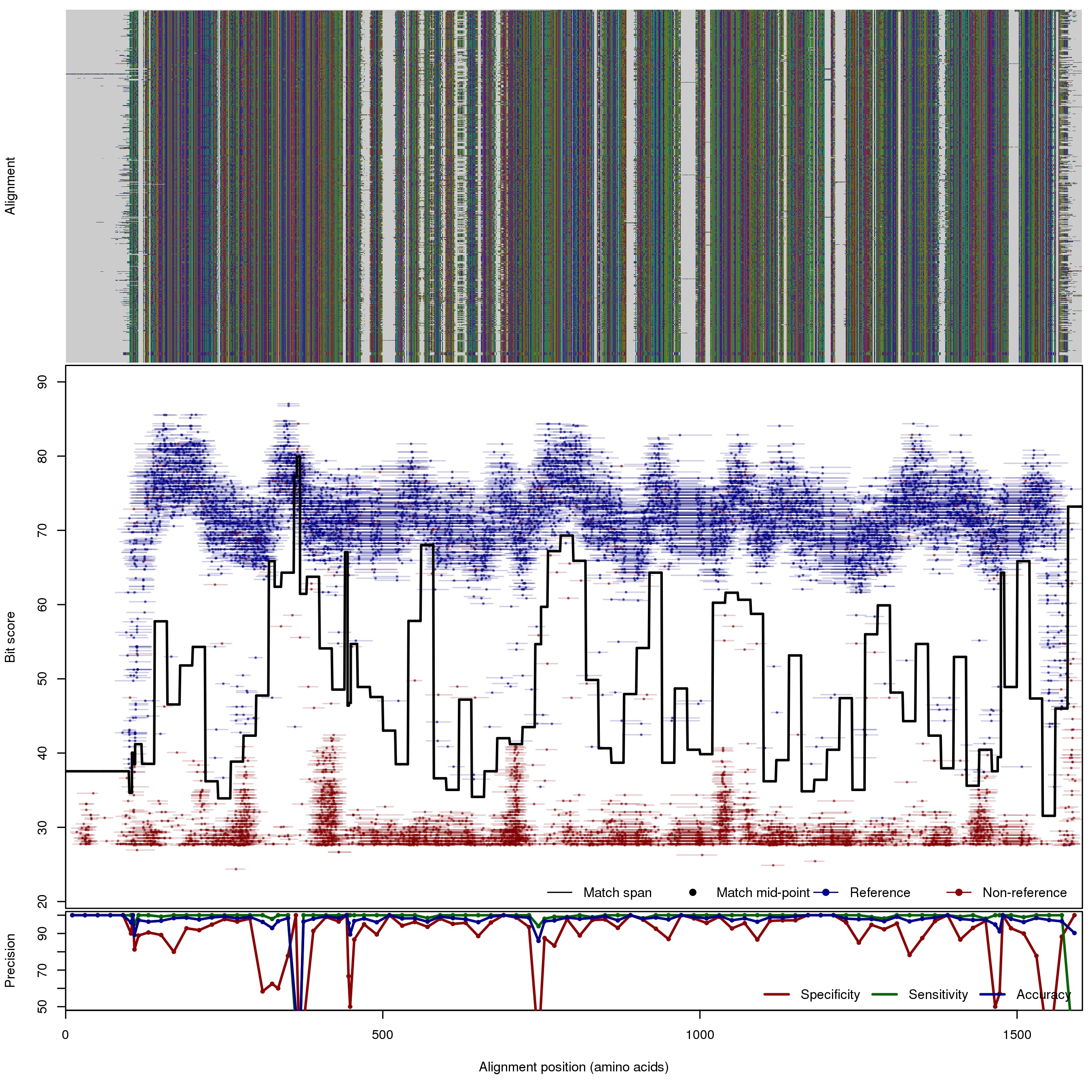 | Download Search online | |||
| 150bp | FNR: 0.27% FPR: 0.2% Acc: 99.8% | 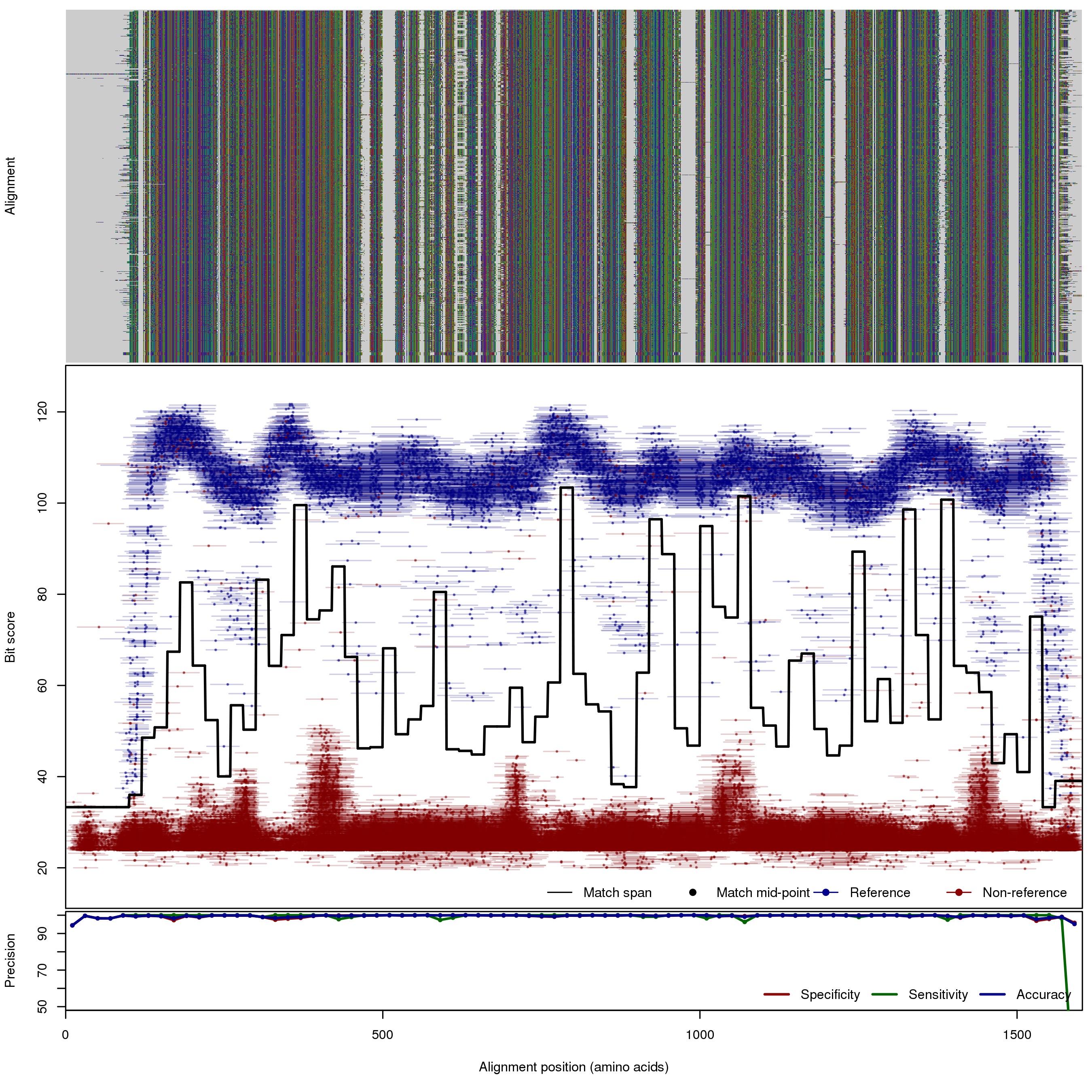 | Download Search online | |||
| 200bp | FNR: 0.24% FPR: 0.12% Acc: 99.87% | 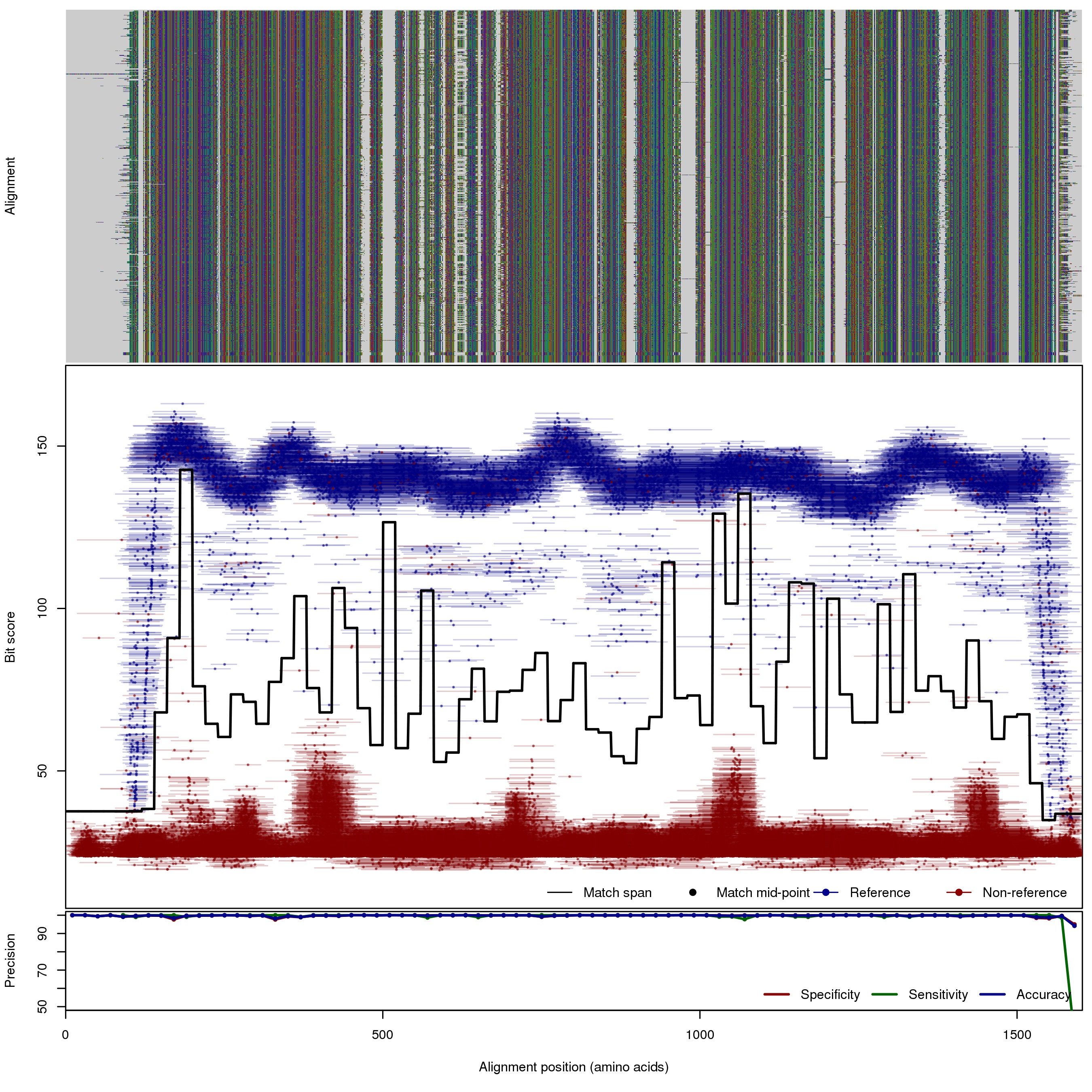 | Download Search online | |||
| 250bp | FNR: 0.39% FPR: 0.09% Acc: 99.91% | 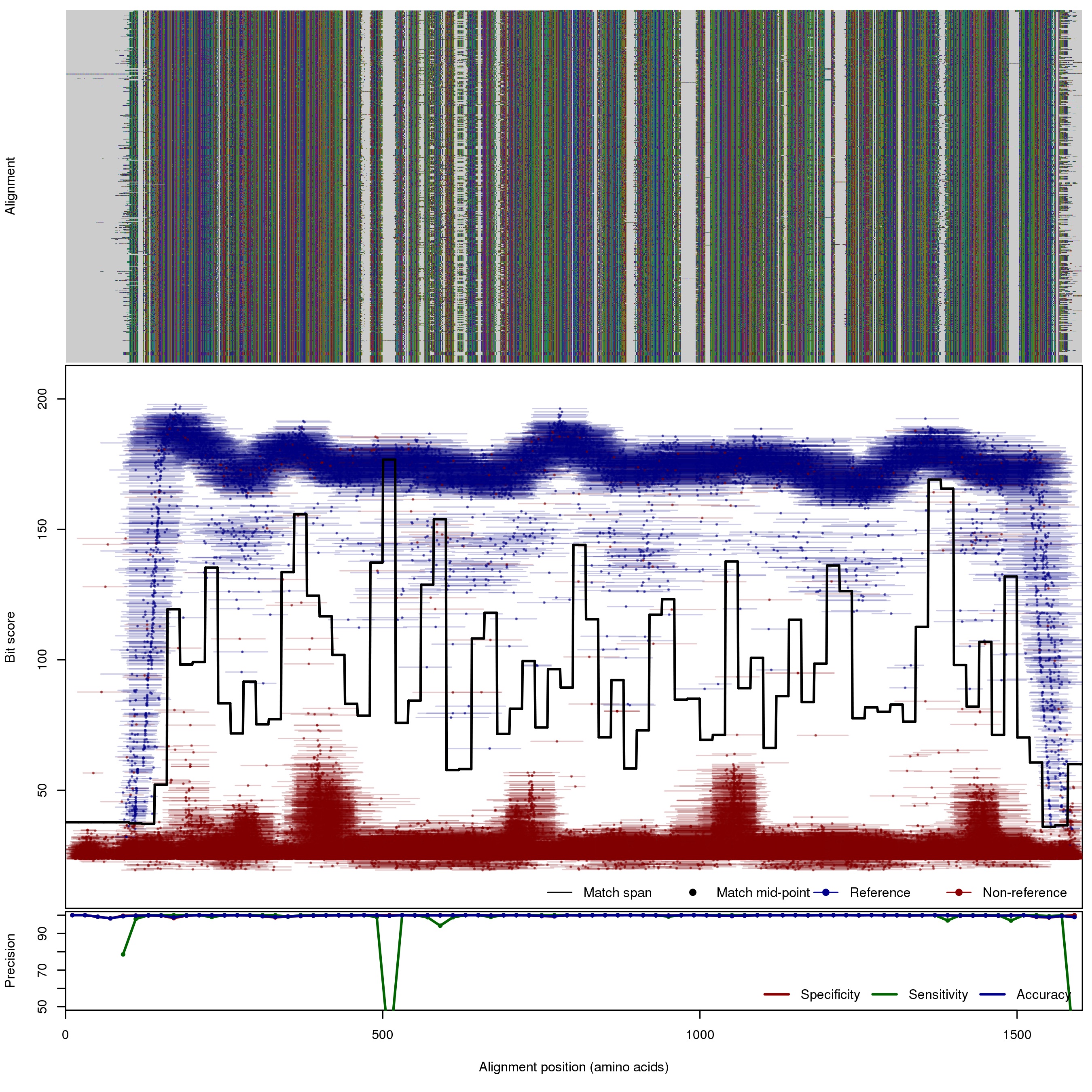 | Download Search online | |||
| 500bp | FNR: 0.49% FPR: 0.04% Acc: 99.95% | 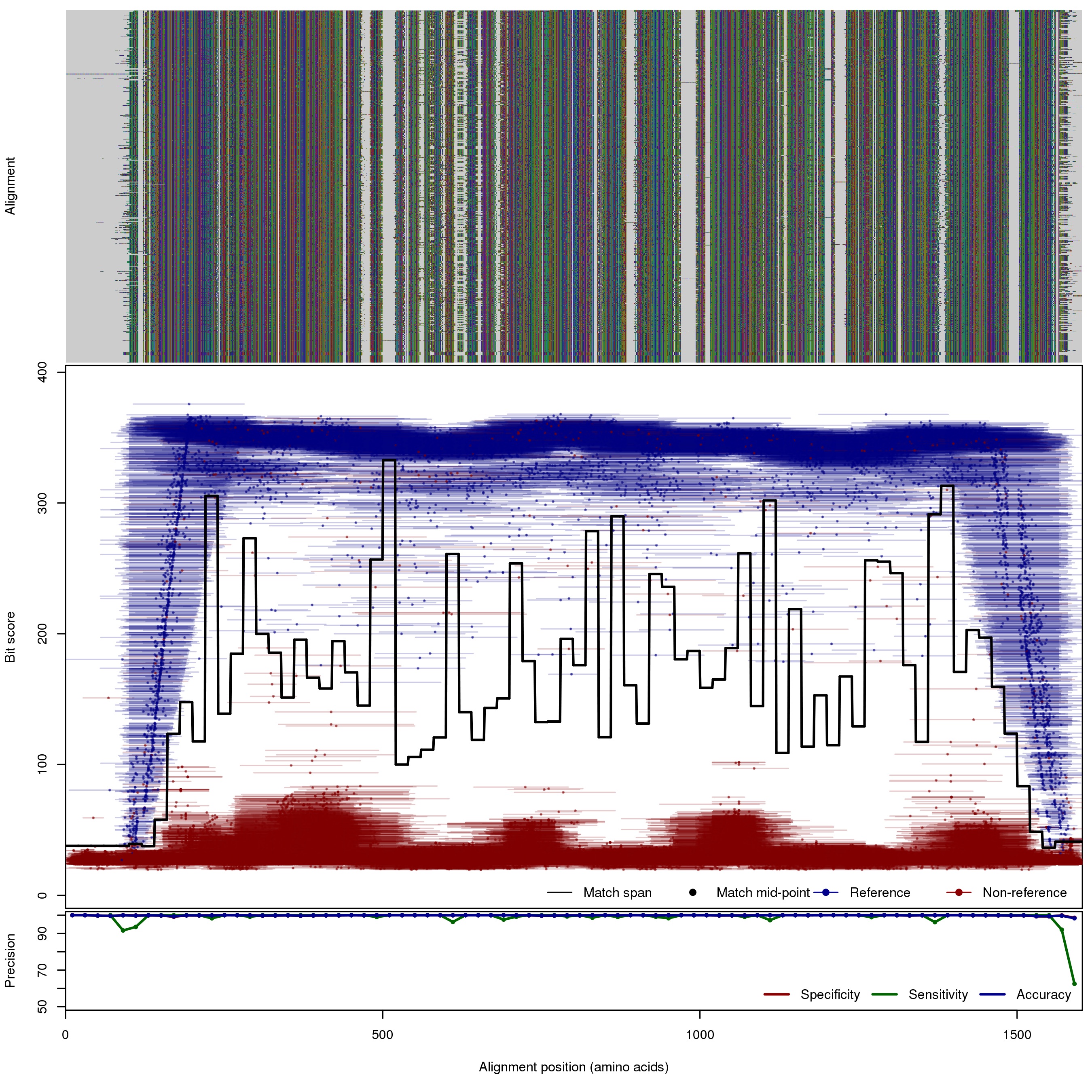 | Download Search online | |||
| NarG_NxrA: Nitrate reductase and nitrite oxidoreductase alpha subunit | ||||||
| NarG_NxrA | 125bp | - | 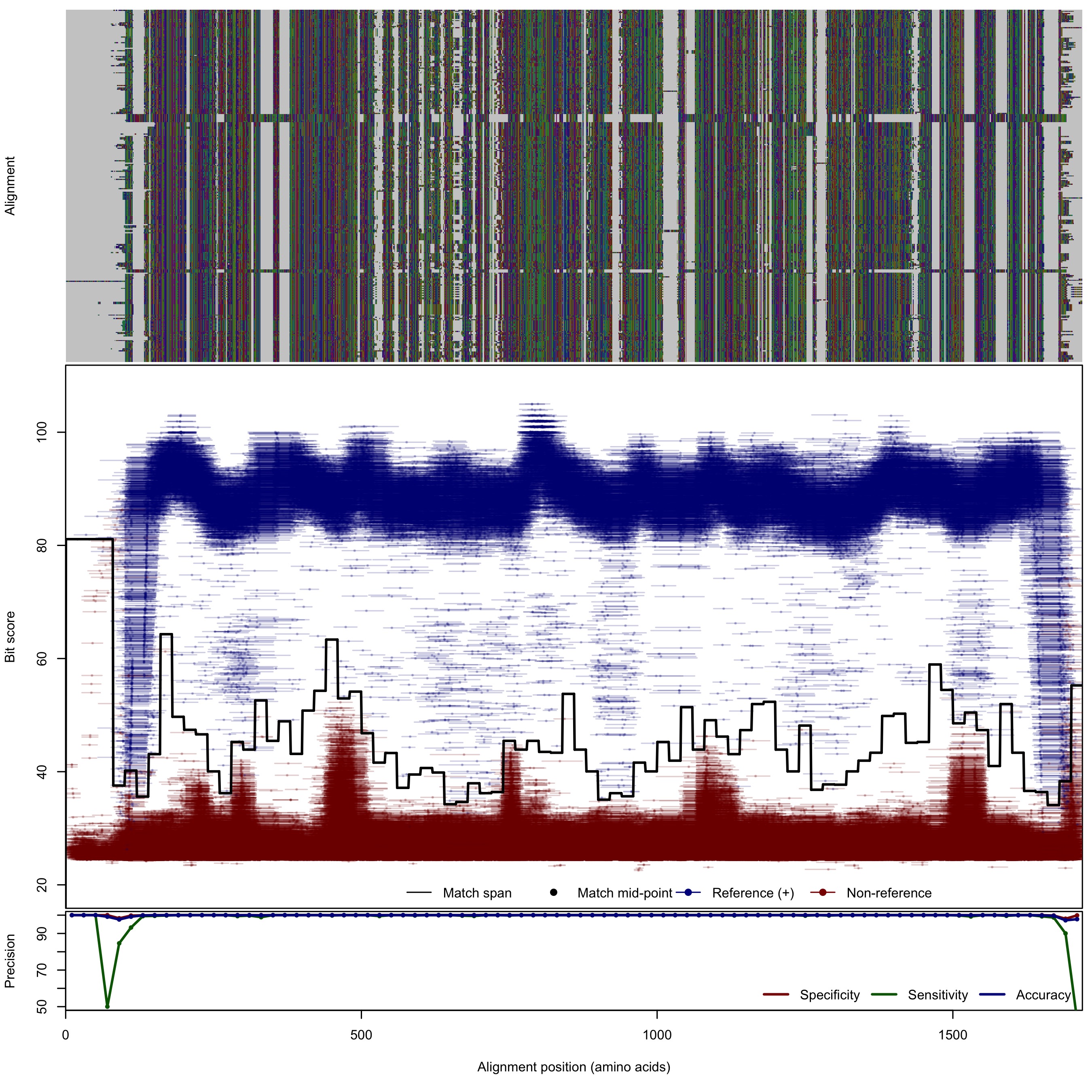 | Download Search online | Positive: 305 proteins Reference sequences | Last update: Apr-18-2017 |
| NifH: Nitrogenase reductase subunit | ||||||
| NifH | 100bp | FNR: 21.7% FPR: 0.0045% Acc: 99.98% |  | Download Search online | Positive: 120 proteins Negative: 56 proteins Reference sequences | Last update: Aug-04-2018 |
| 150bp | FNR: 23.09% FPR: 0.0031% Acc: 99.98% | 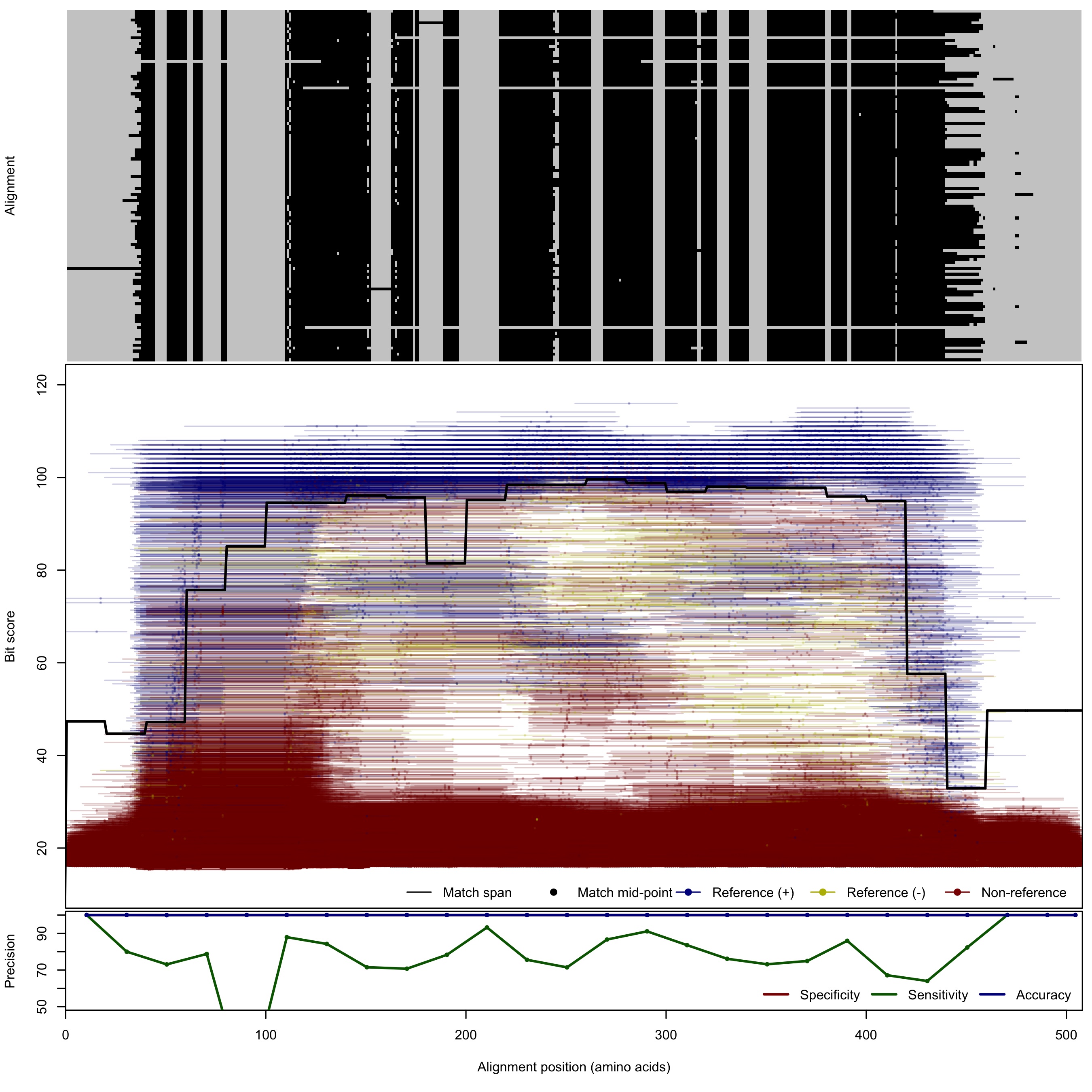 | Download Search online | |||
| 200bp | FNR: 27.1% FPR: 0.0036% Acc: 99.98% | 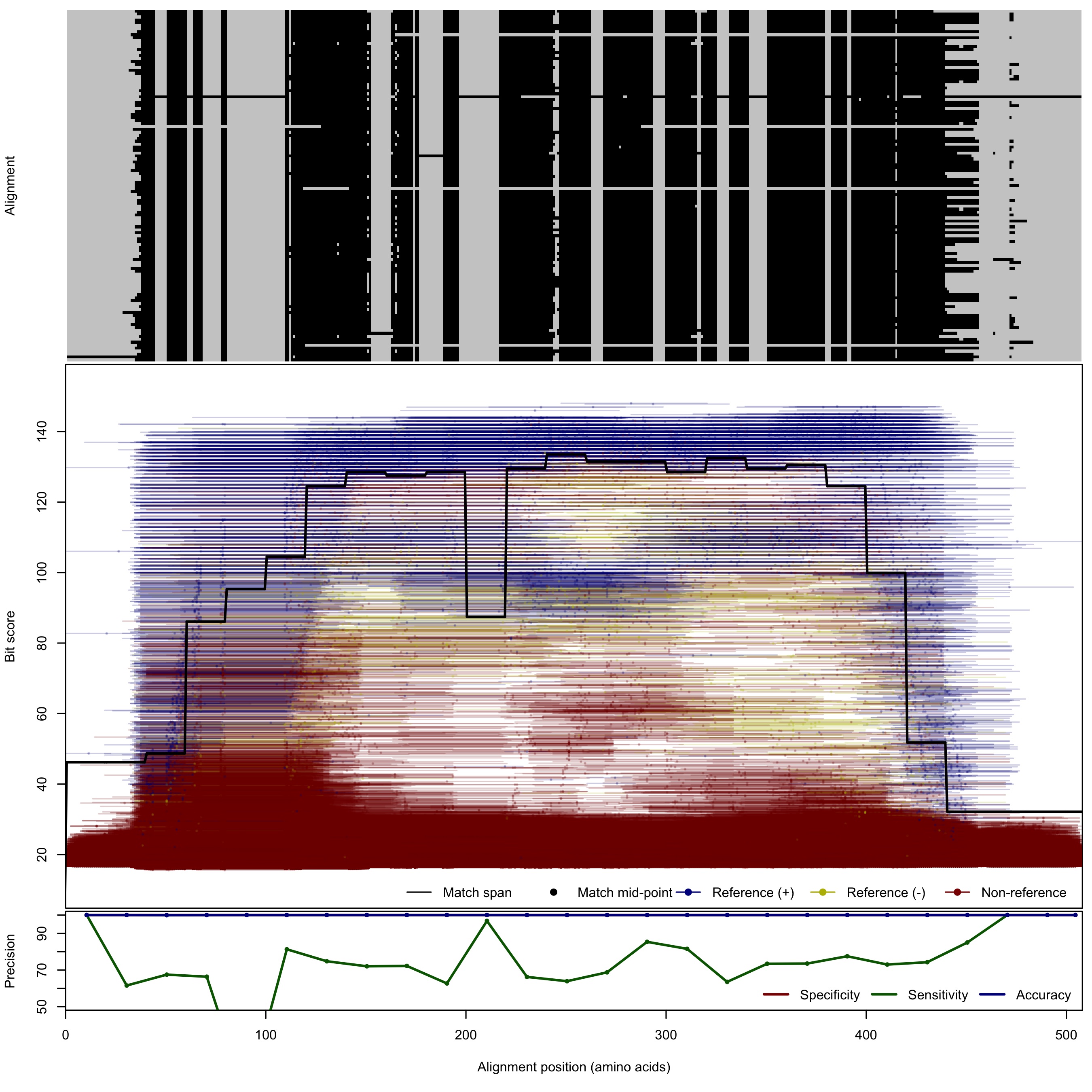 | Download Search online | |||
| NirK: Nitrite reductase | ||||||
| NirK | 80bp | FNR: 1.99% FPR: 2.72% Acc: 97.82% | 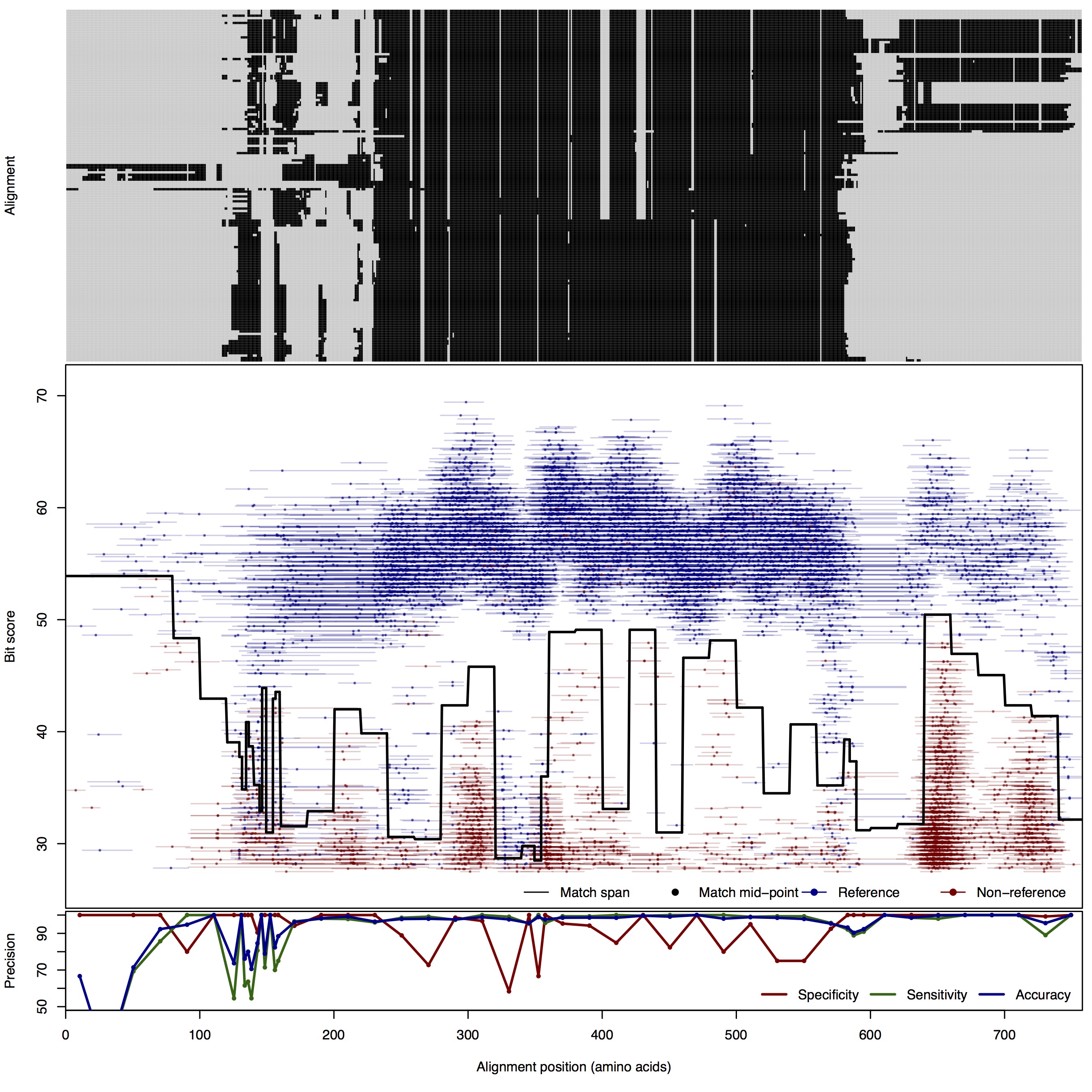 | Download Search online | Positive: 147 proteins Reference sequences | Last update: Jul-14-2015 |
| 100bp | FNR: 2.76% FPR: 1.49% Acc: 97.71% | 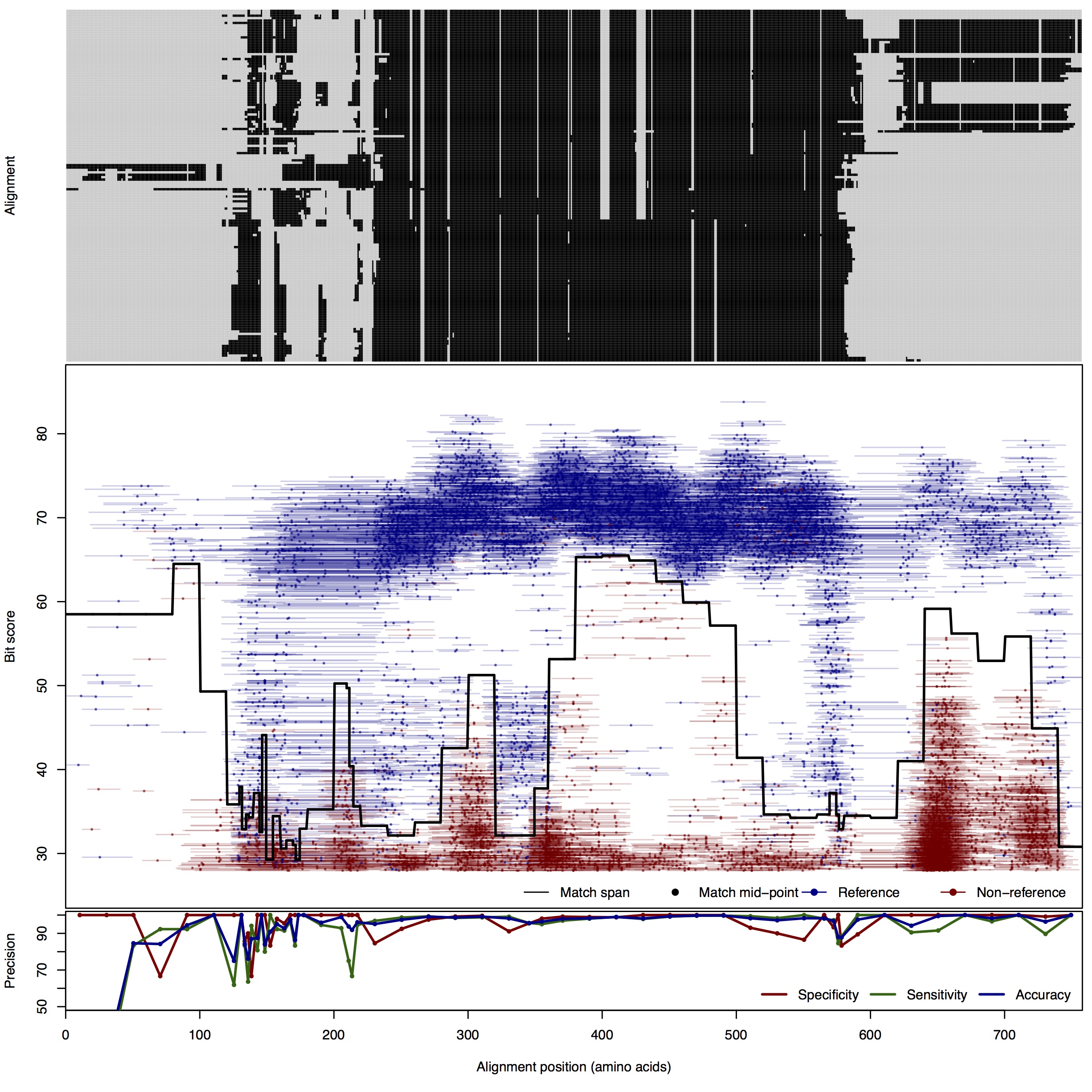 | Download Search online | |||
| 150bp | FNR: 2.17% FPR: 0.73% Acc: 98.63% | 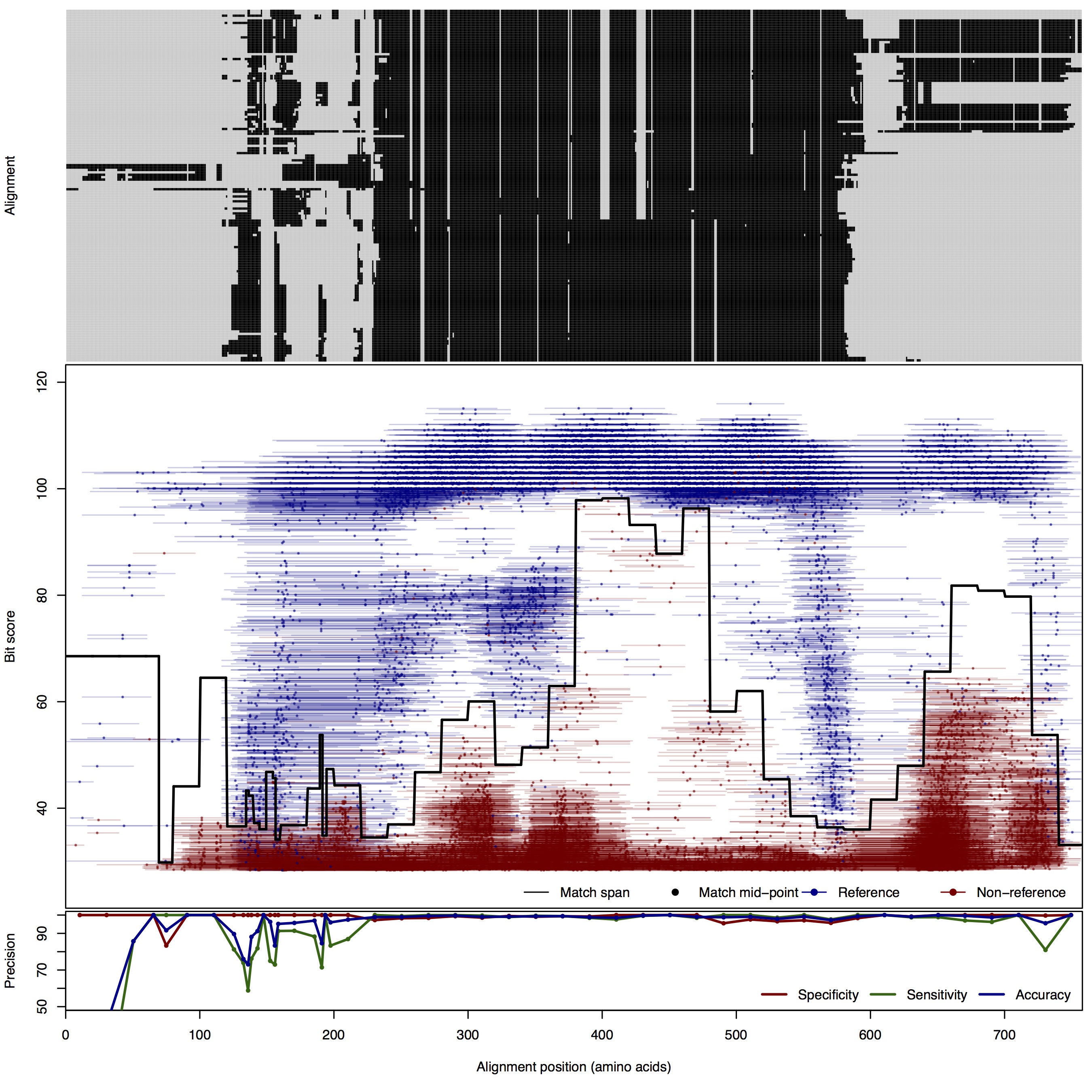 | Download Search online | |||
| 200bp | FNR: 1.72% FPR: 0.57% Acc: 99.02% | 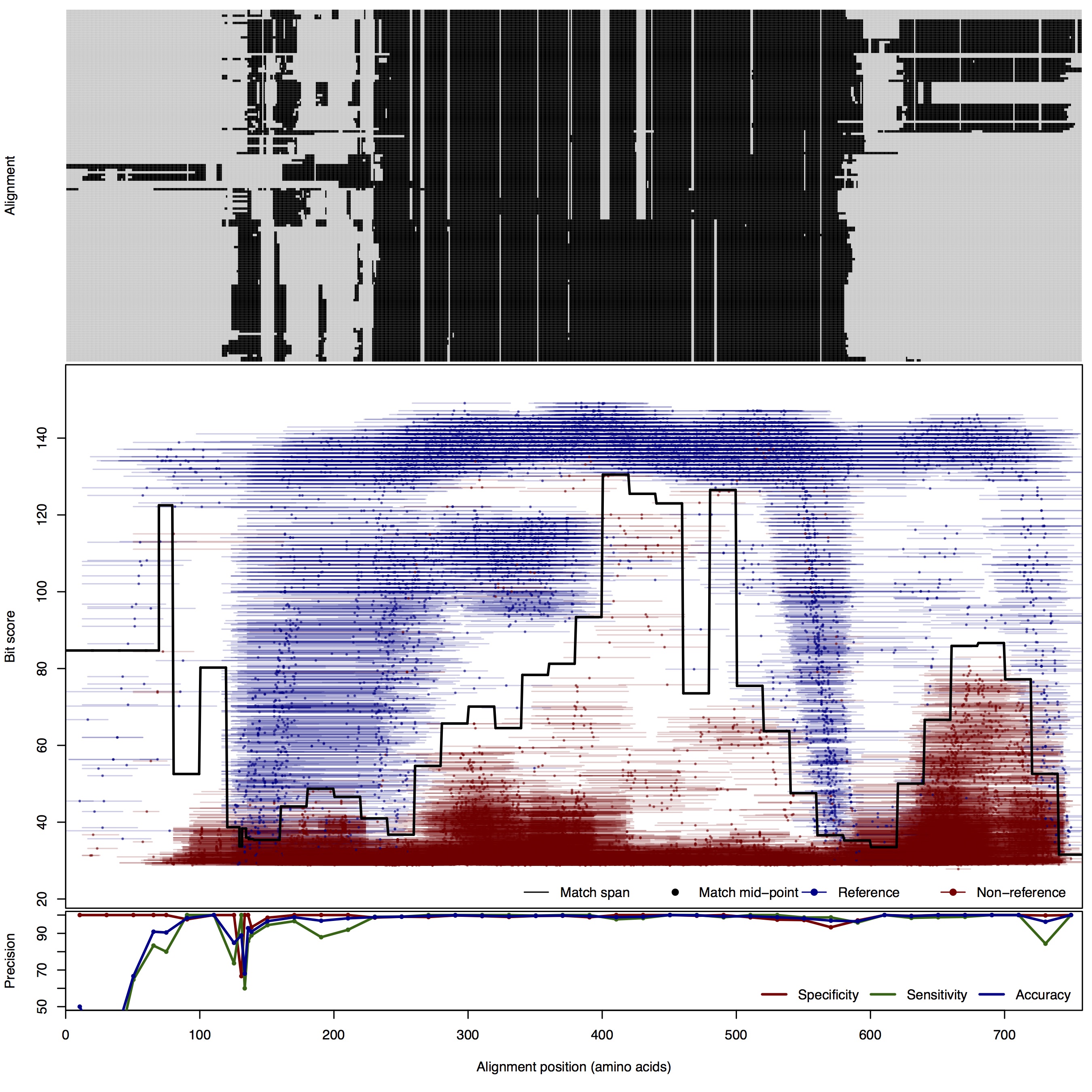 | Download Search online | |||
| 250bp | FNR: 1.93% FPR: 0.49% Acc: 99.06% | 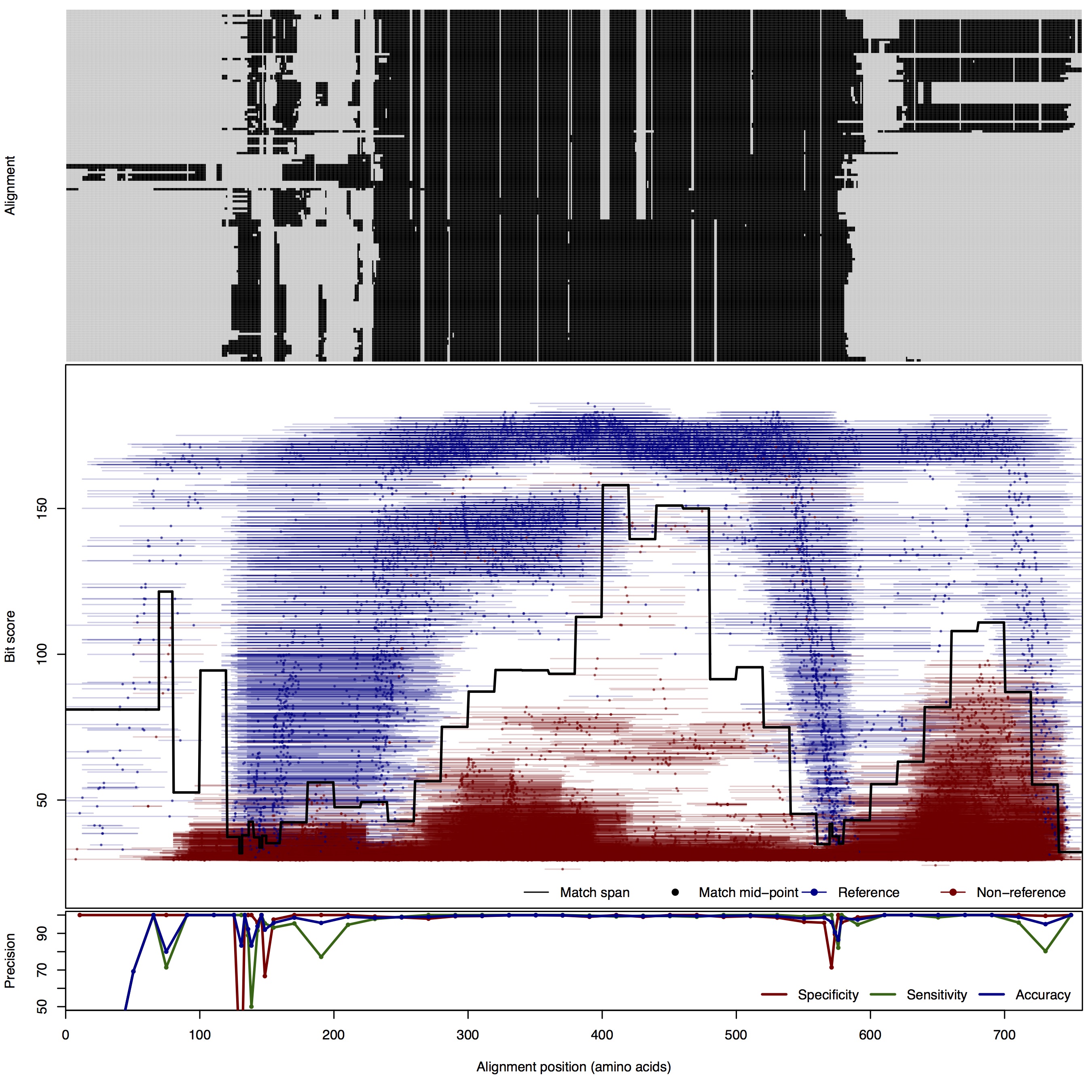 | Download Search online | |||
| 500bp | FNR: 1.64% FPR: 0.68% Acc: 99.04% | 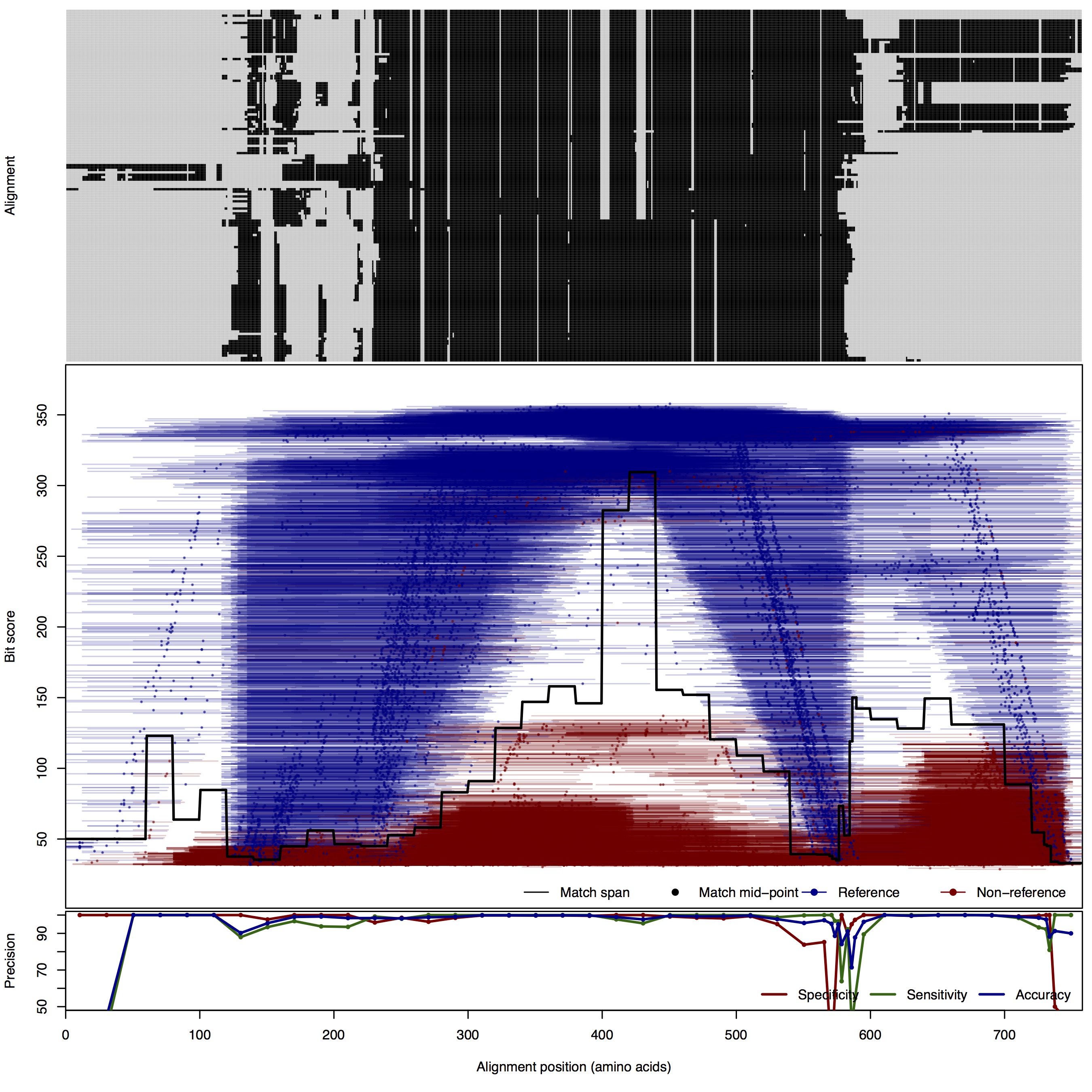 | Download Search online | |||
| NirK_cladeThau | 125bp | - | 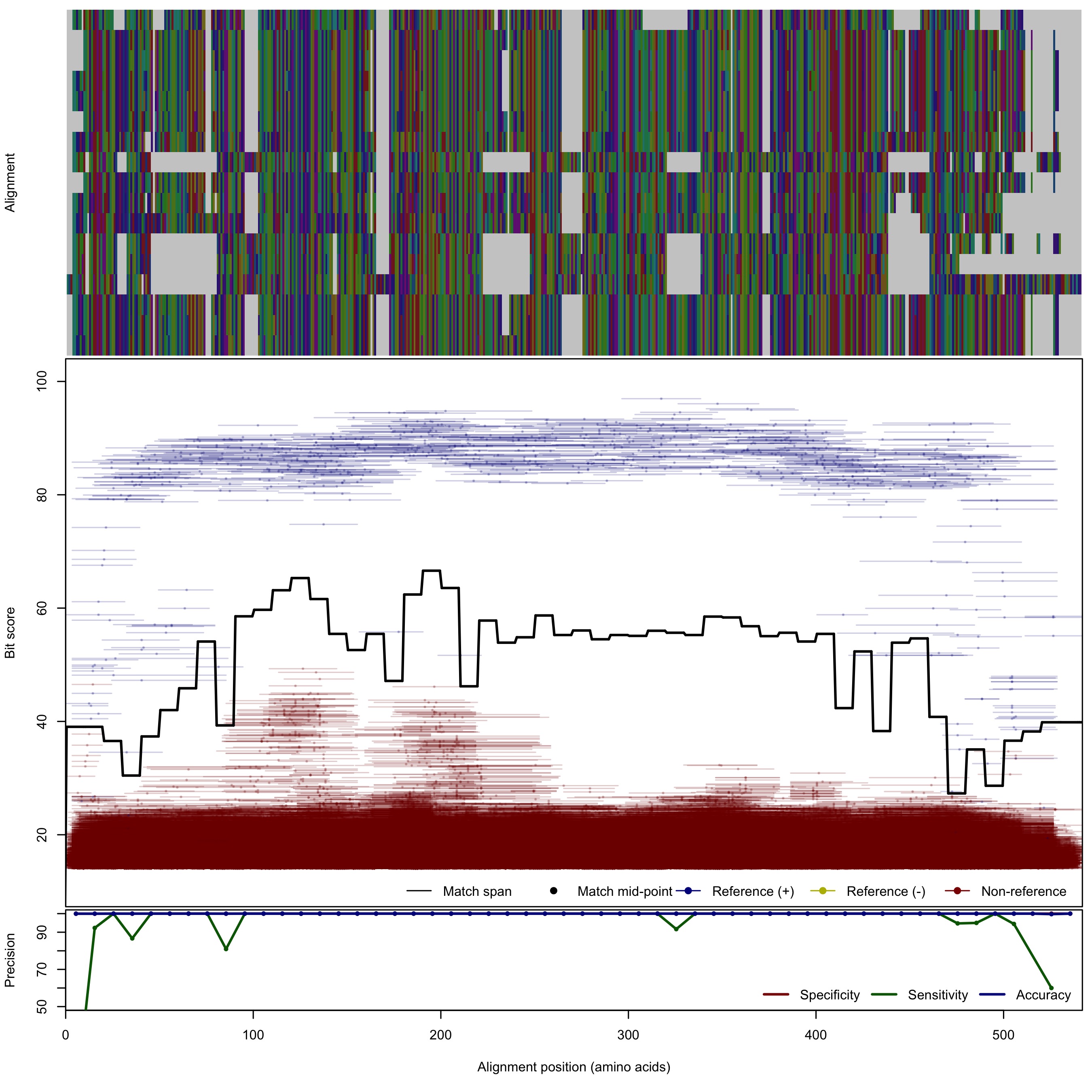 | Download Search online | Positive: 18 proteins Reference sequences | Last update: Apr-18-2017 |
| NirK_cladeIII | 125bp | - | 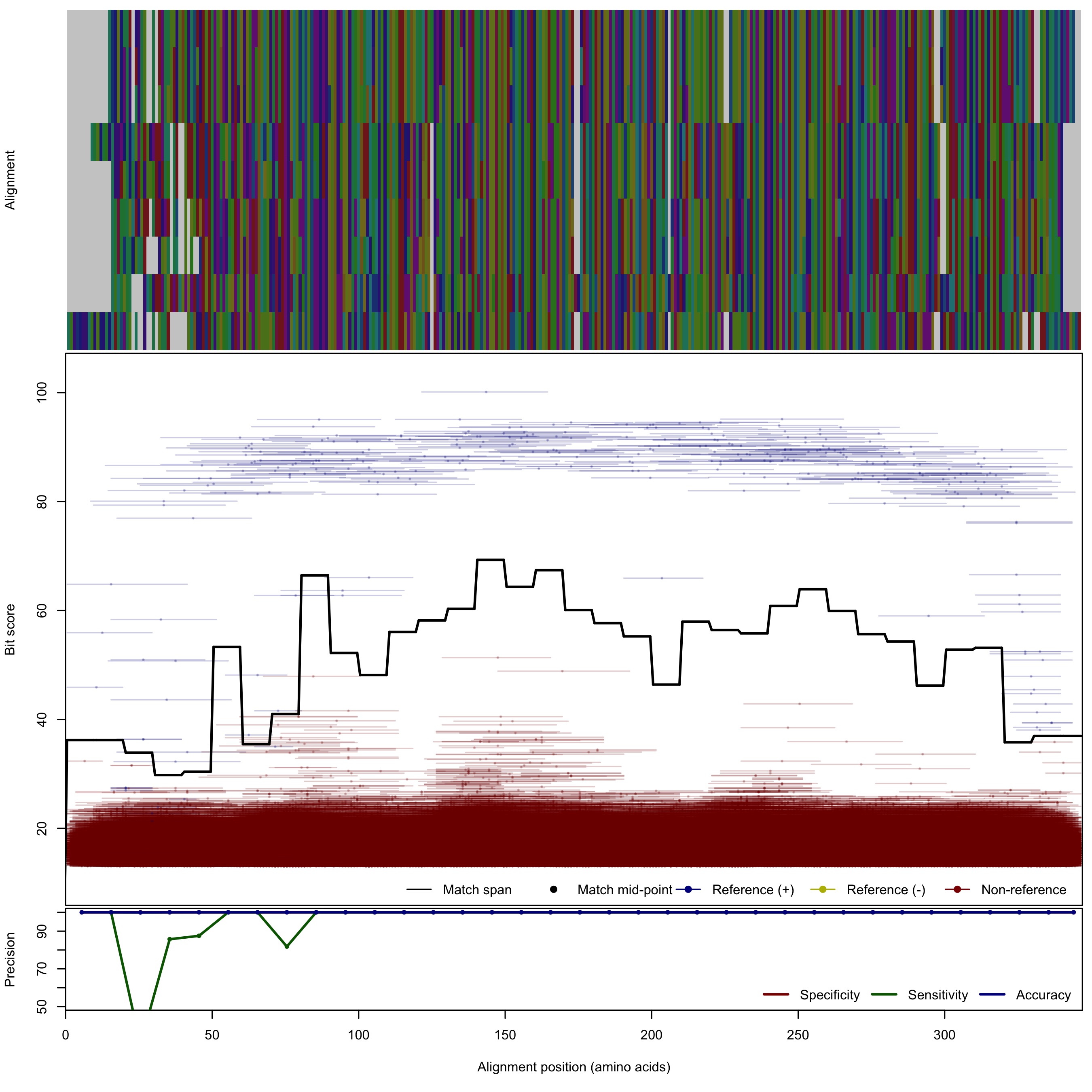 | Download Search online | Positive: 10 proteins Reference sequences | Last update: Apr-18-2017 |
| NirK_cladeI-II | 125bp | - | 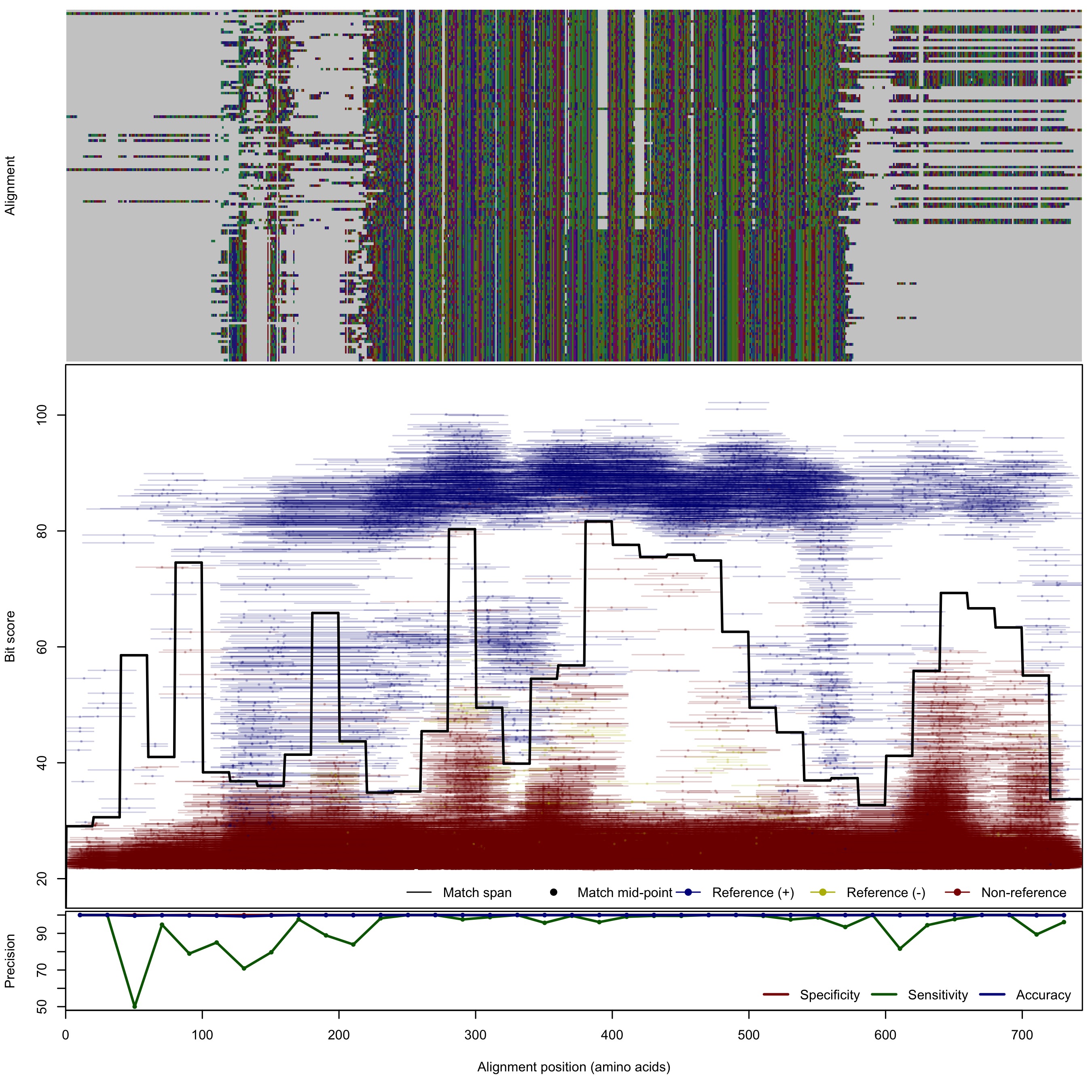 | Download Search online | Positive: 134 proteins Negative: 8 proteins Reference sequences | Last update: Apr-18-2017 |
| NorB: Nitric oxide reductase | ||||||
| NorB | 125bp | - | 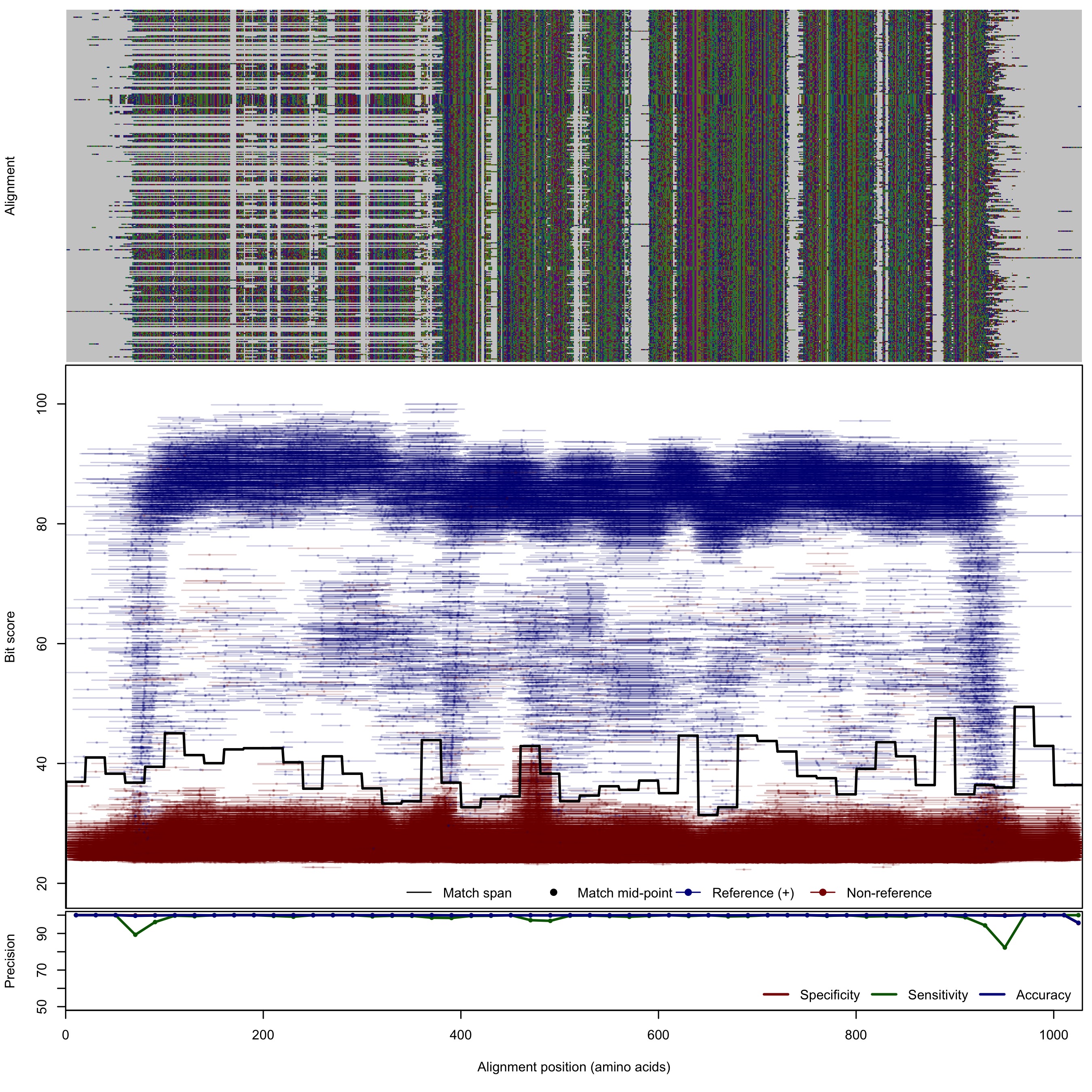 | Download Search online | Positive: 304 proteins Reference sequences | Last update: Apr-18-2017 |
| NosZ: Nitrous oxide reductase | ||||||
| NosZ | 80bp | FNR: 0.89% FPR: 7.44% Acc: 98.65% | 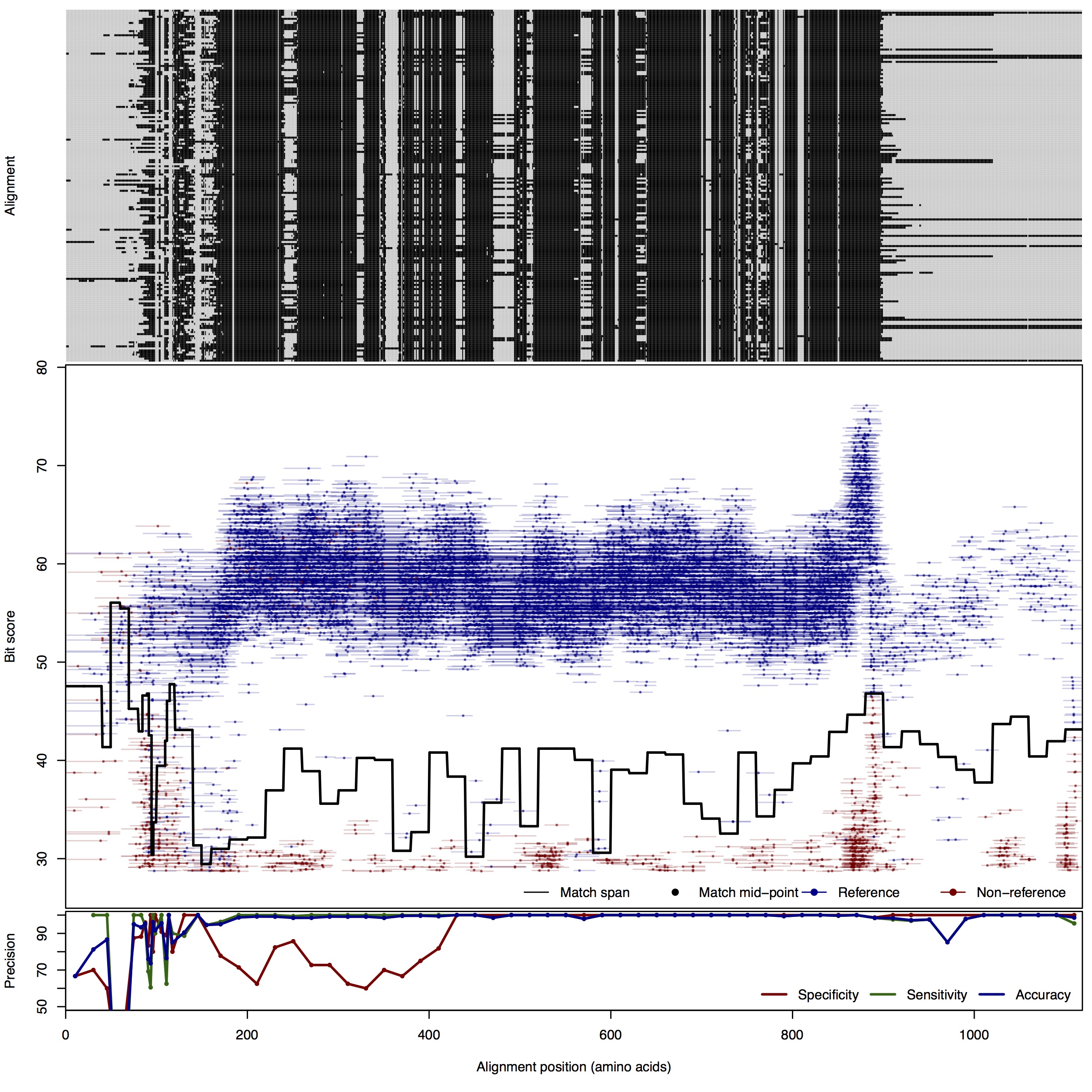 | Download Search online | Positive: 173 proteins Reference sequences | Last update: Jul-14-2015 |
| 100bp | FNR: 1.09% FPR: 5.44% Acc: 98.33% | 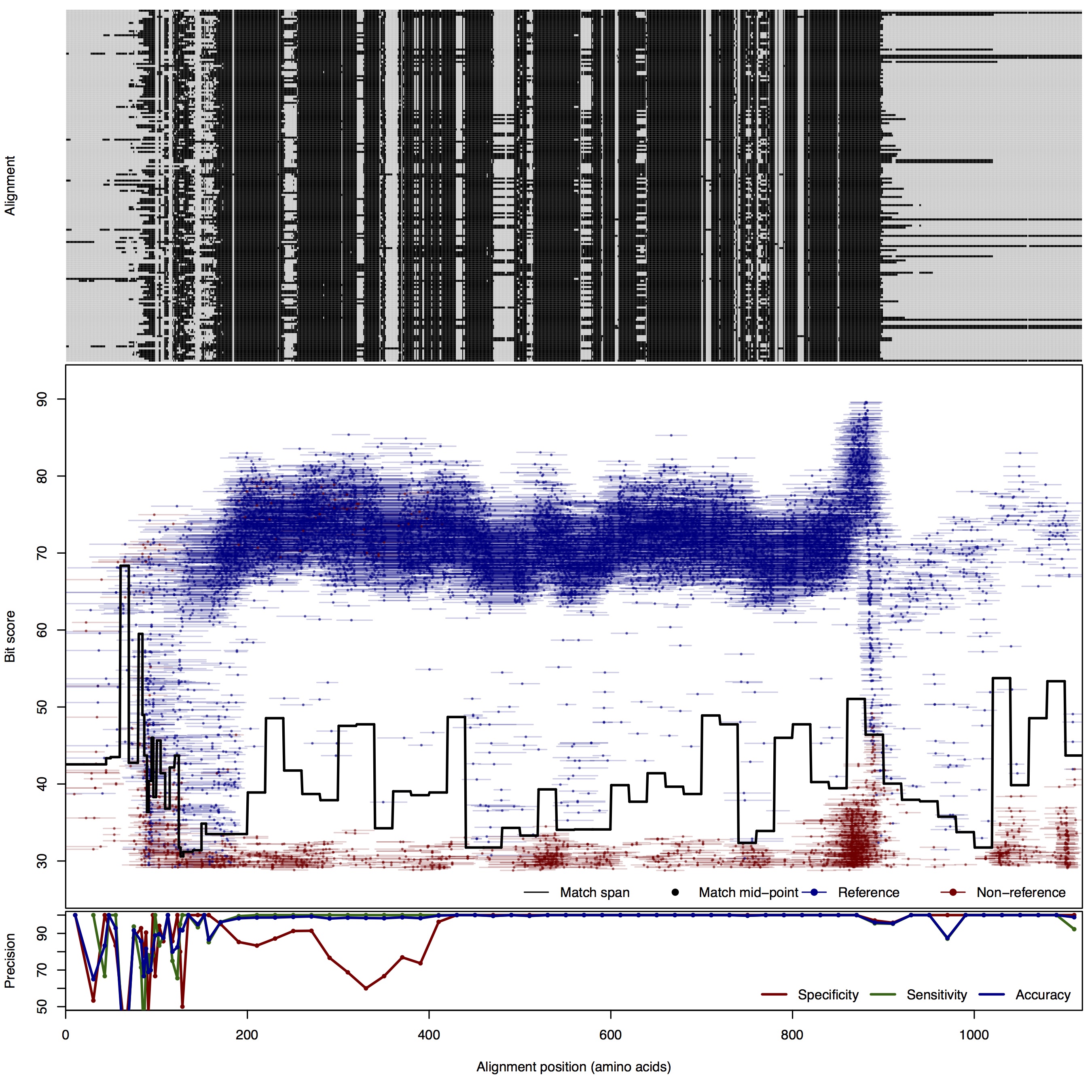 | Download Search online | |||
| 125bp | FNR: 1.09% FPR: 0.66% Acc: 99.01% | 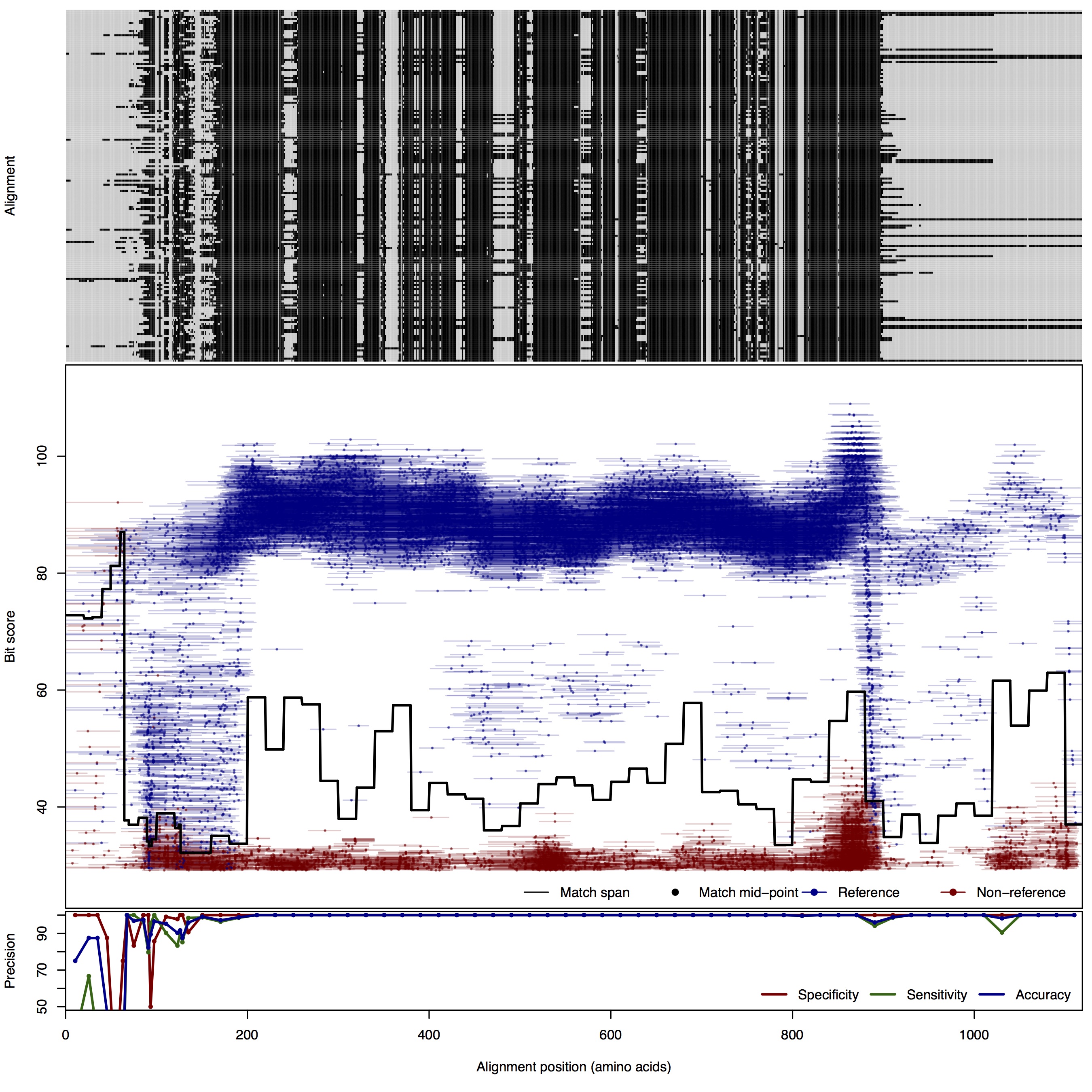 | Download Search online | |||
| 150bp | FNR: 0.73% FPR: 1.82% Acc: 98.94% | 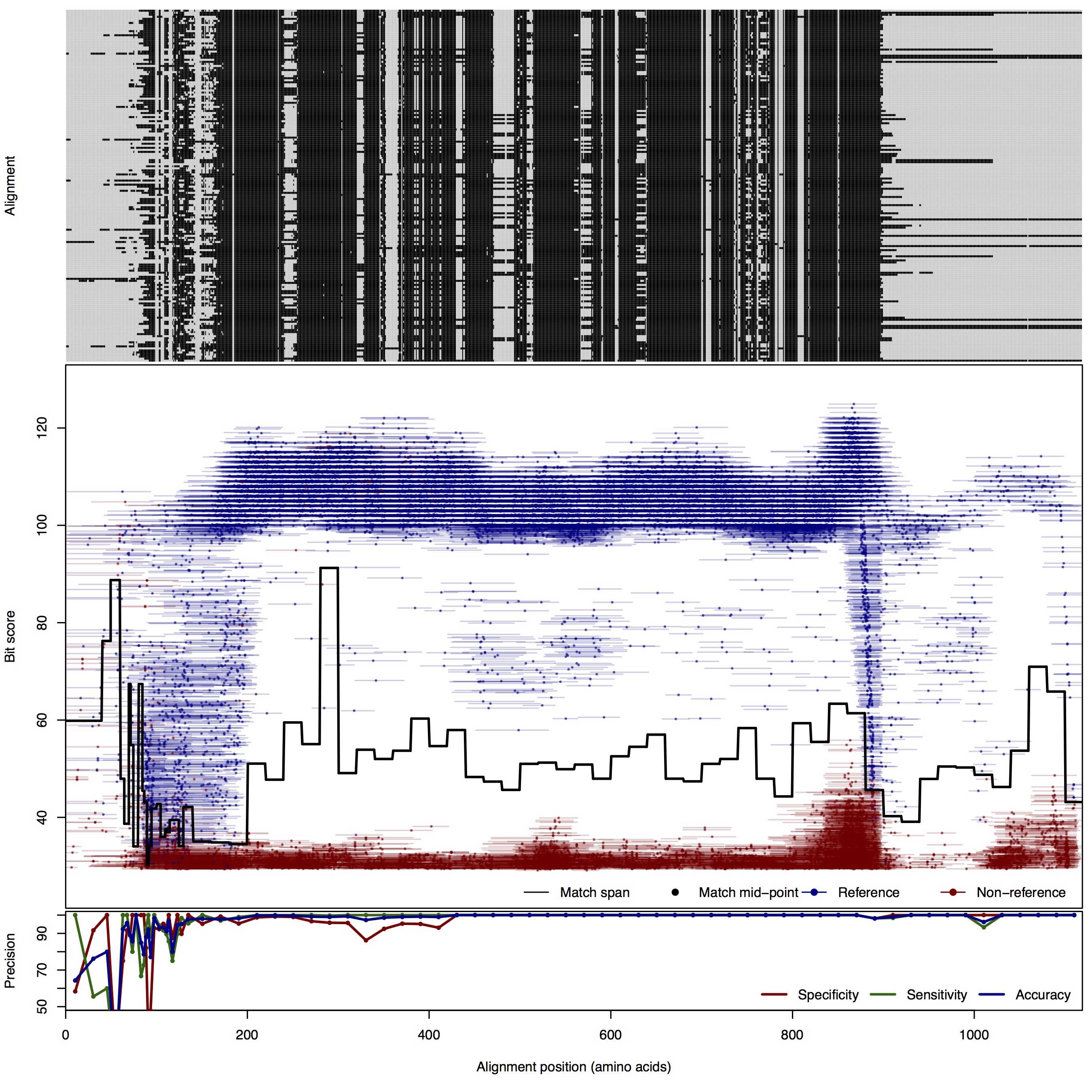 | Download Search online | |||
| 200bp | FNR: 1.16% FPR: 0.84% Acc: 98.97% | 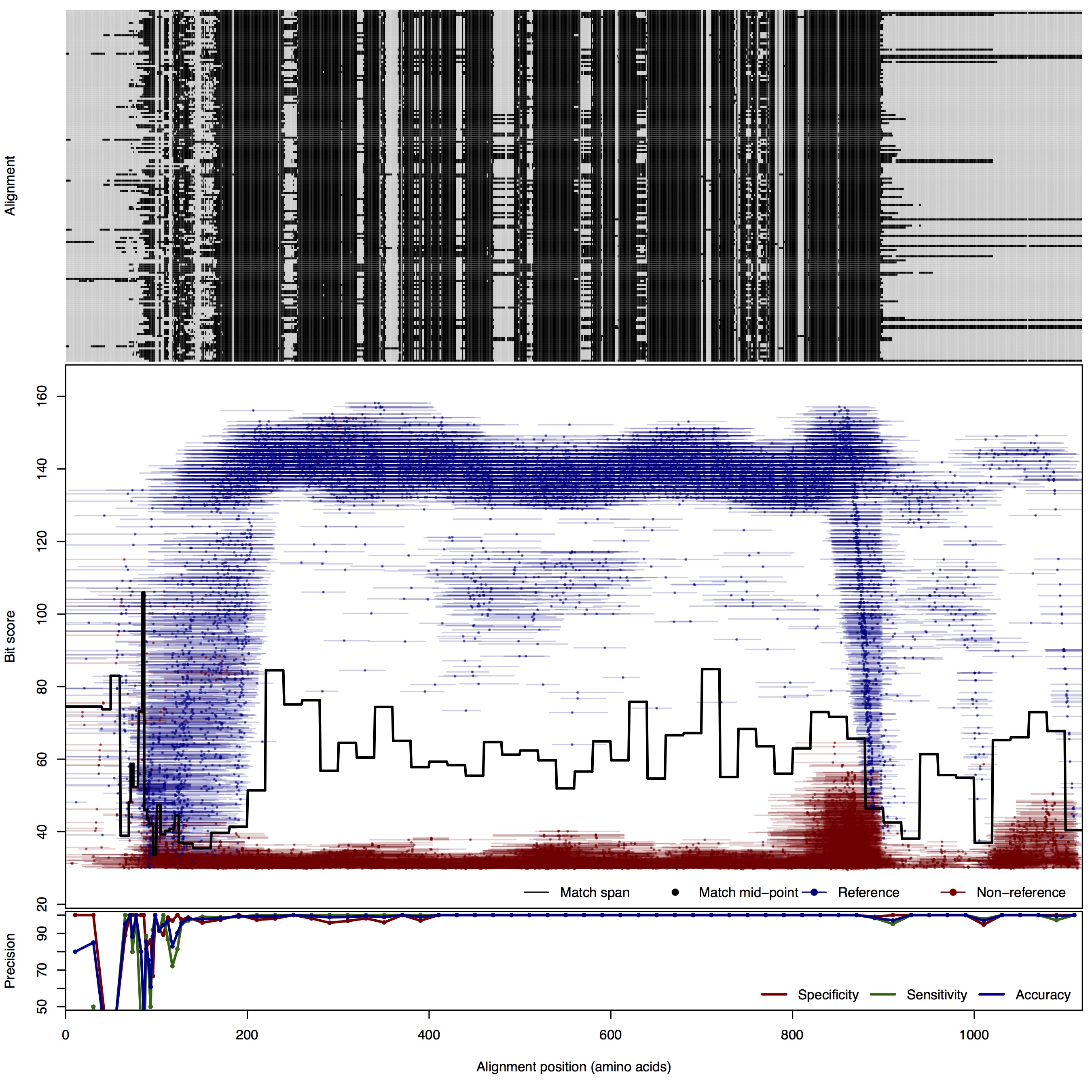 | Download Search online | |||
| 250bp | FNR: 0.61% FPR: 0.75% Acc: 99.33% | 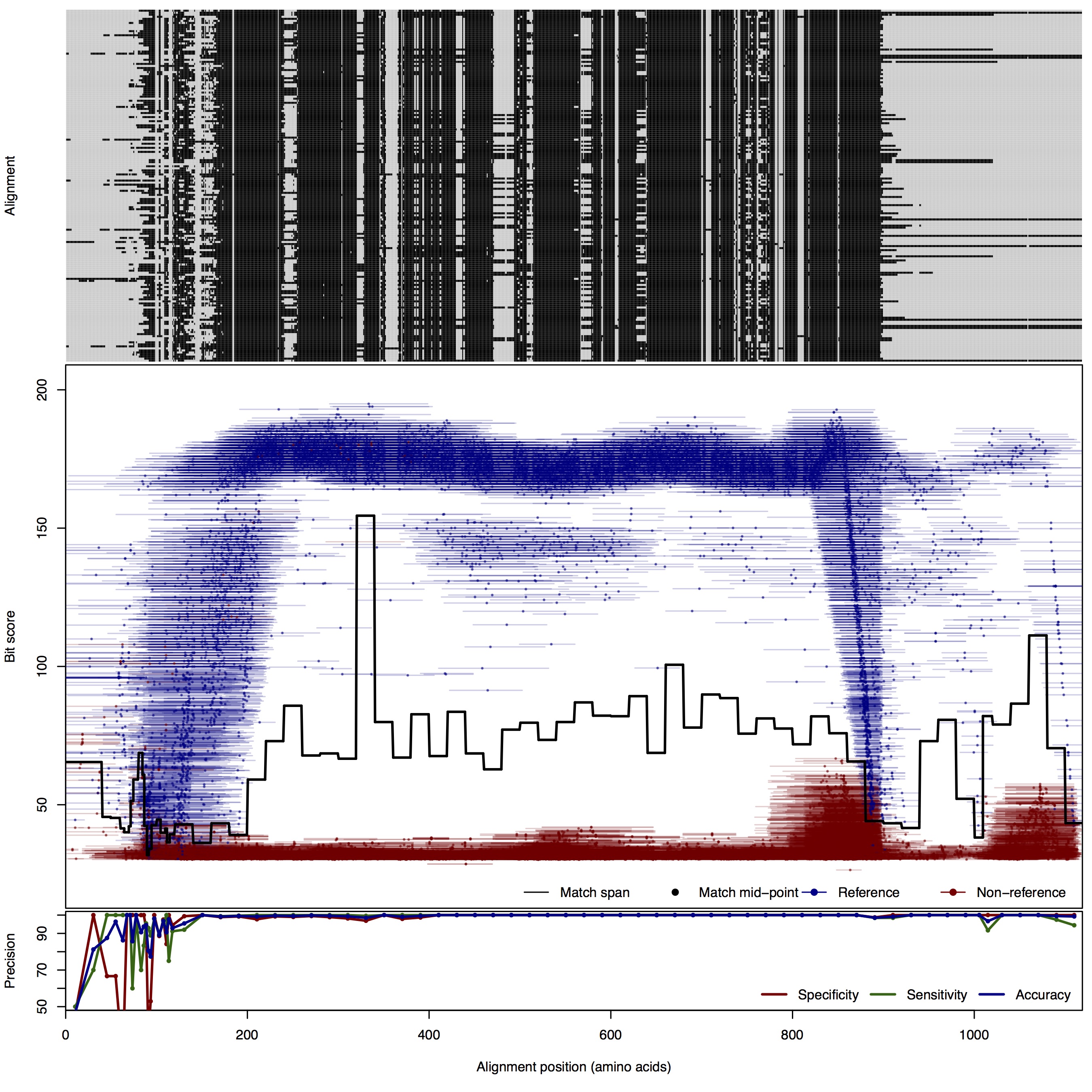 | Download Search online | |||
| 500bp | FNR: 0.73% FPR: 0.24% Acc: 99.51% | 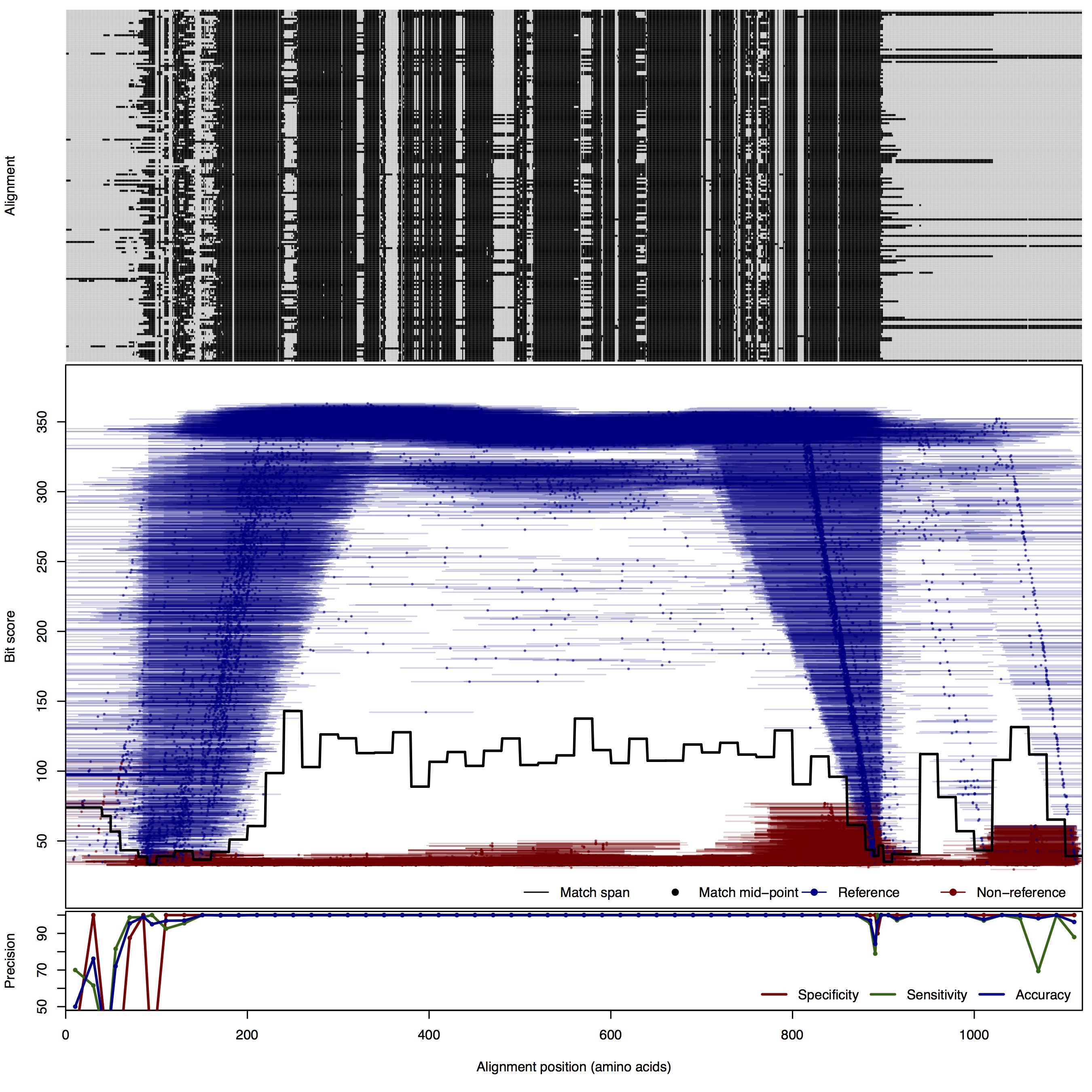 | Download Search online | |||
| NosZ_v2 | 125bp | - | 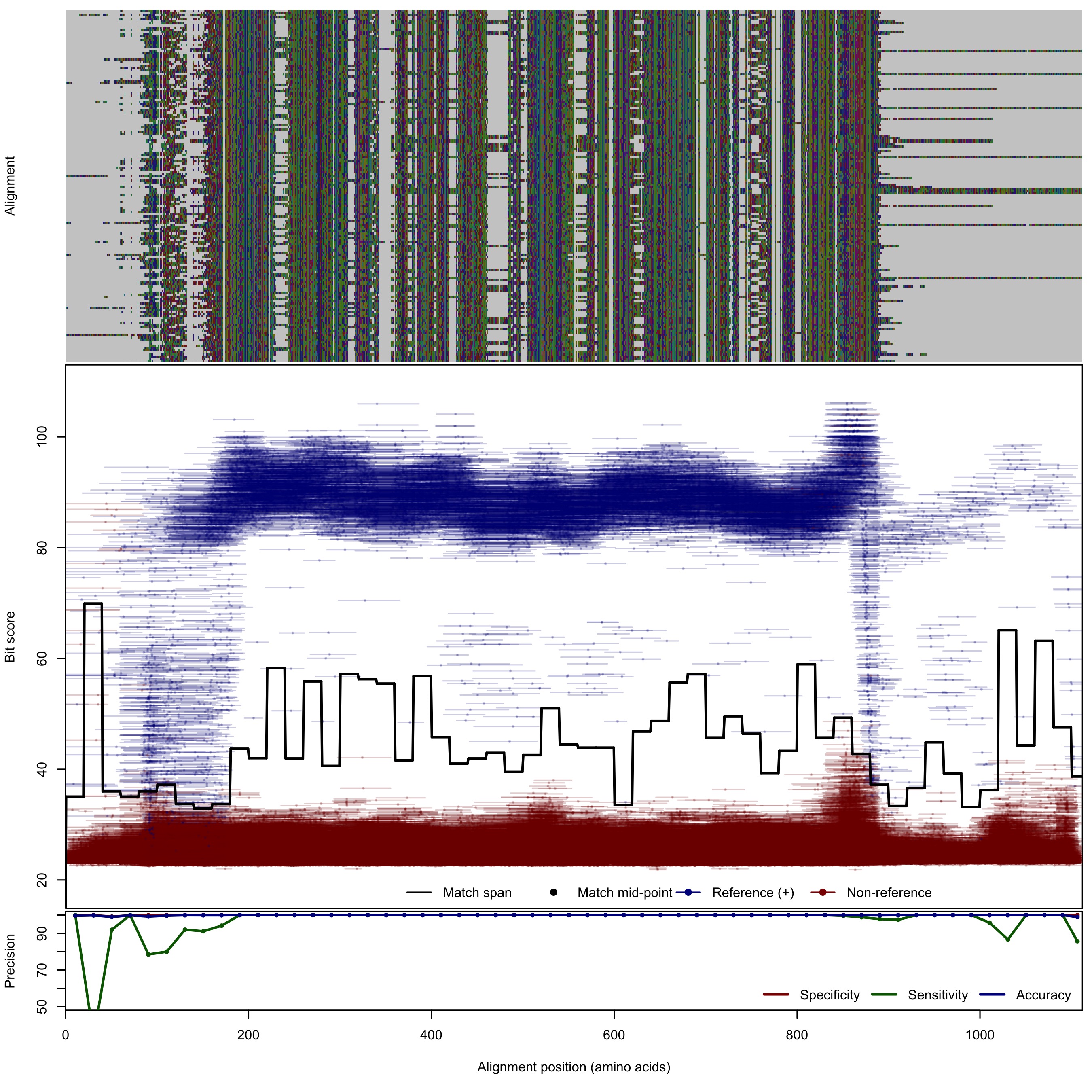 | Download Search online | Positive: 167 proteins Reference sequences | Last update: Apr-18-2017 |
| NrfA: Nitrite reductase, formate-dependent | ||||||
| NrfA | 125bp | - | 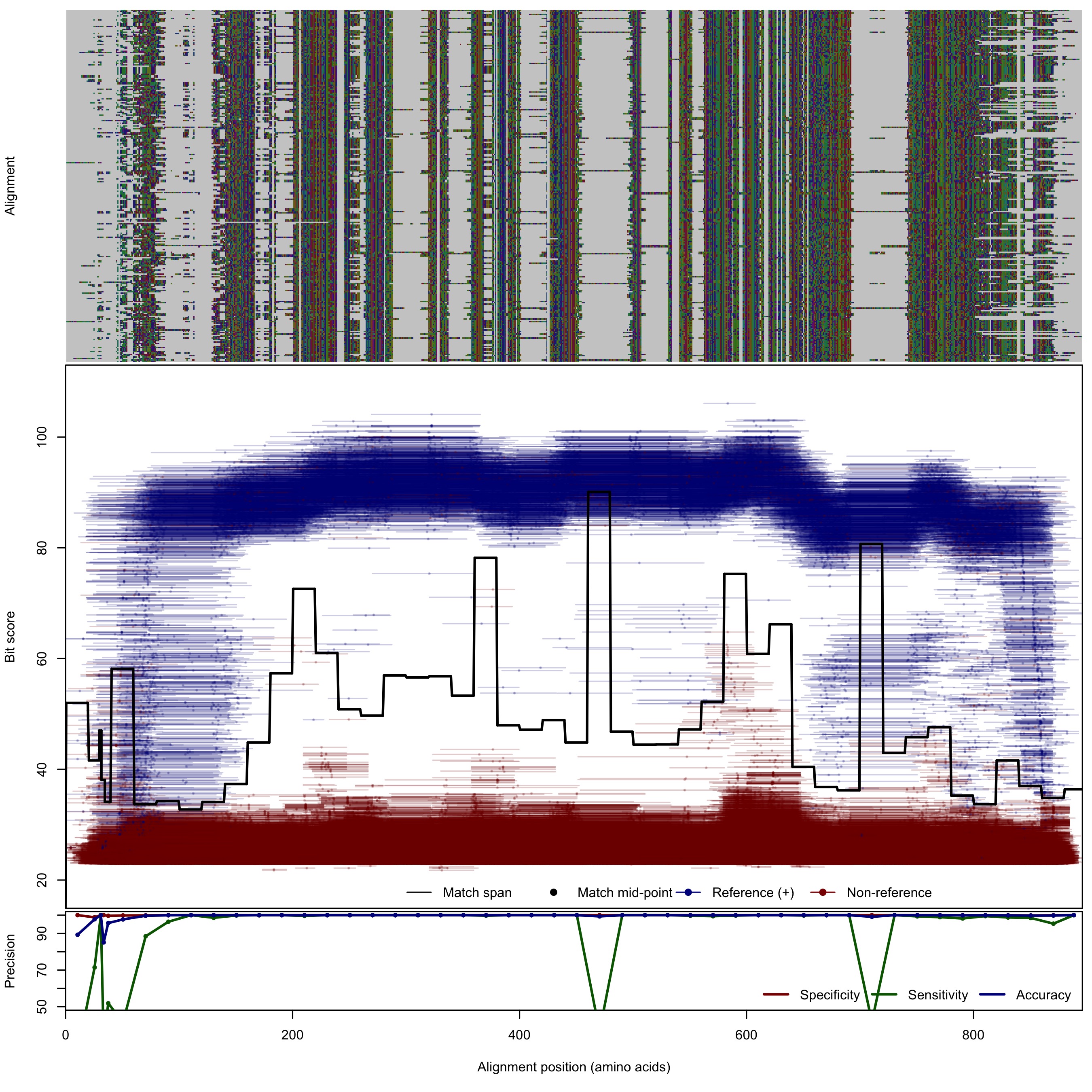 | Download Search online | Positive: 260 proteins Negative: 8 proteins Reference sequences | Last update: Apr-18-2017 |
| OxyT: N,N-dimethyltransferase | ||||||
| OxyT | 150bp | - | 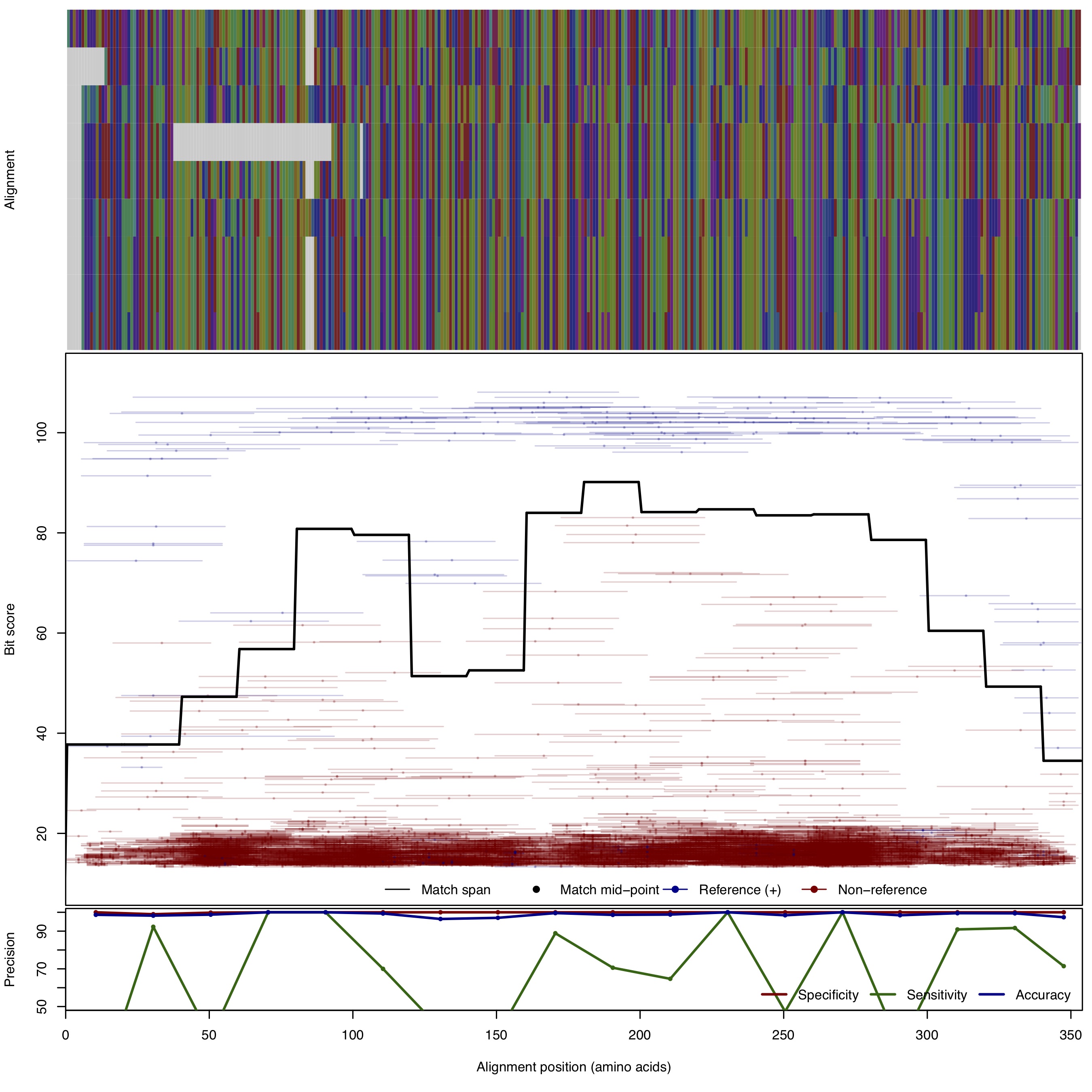 | Download Search online | Positive: 10 proteins Negative: 4 proteins Reference sequences | Tetracycline biosynthesis Last update: Apr-13-2018 |
| RpoB: Ribosomal RNA polymerase B subunit | ||||||
| RpoB | 80bp | FNR: 0.05% FPR: 0.64% Acc: 99.9% | 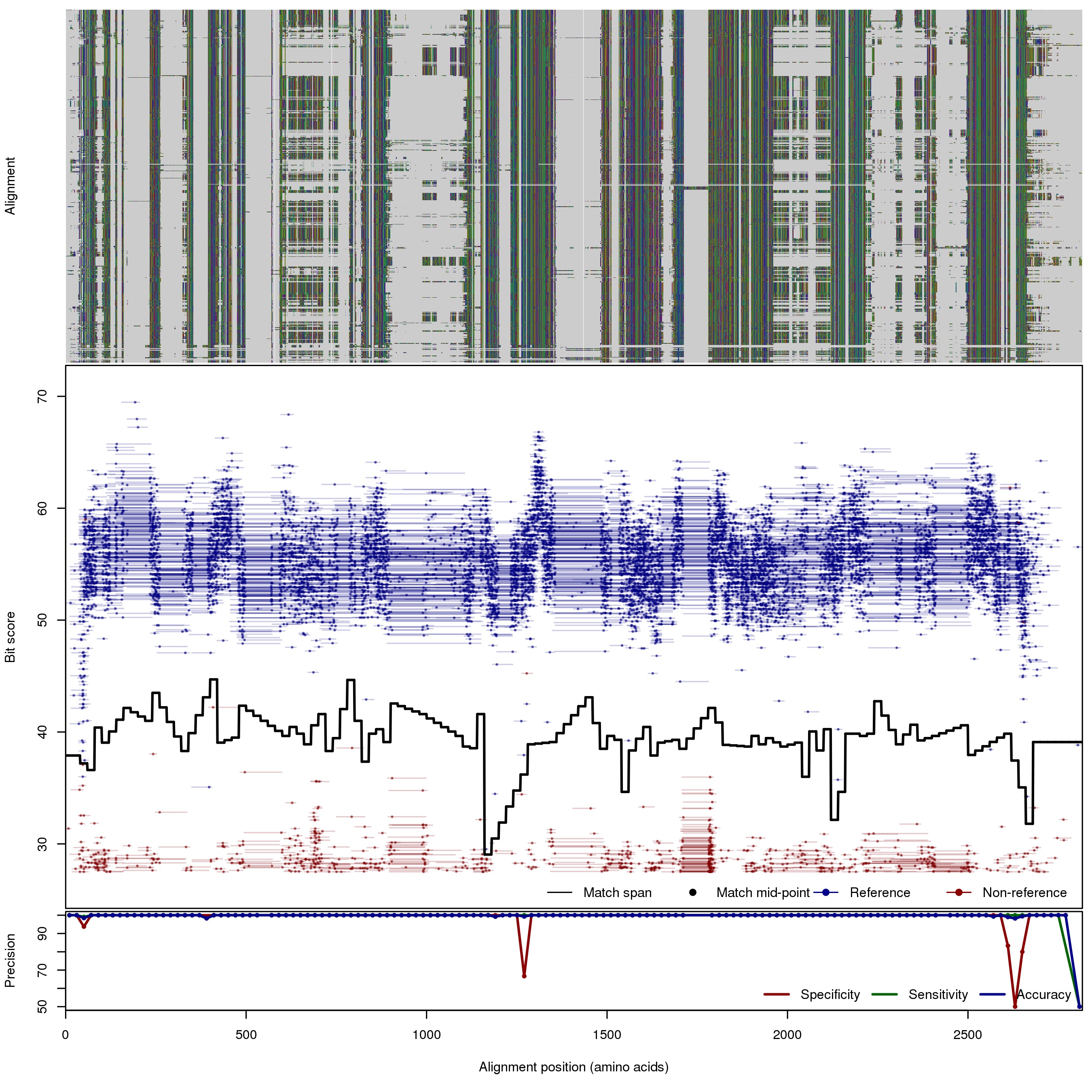 | Download Search online | Positive: 757 proteins Reference sequences | [with --search diamond]Last update: Jun-09-2015 |
| 100bp | FNR: 0.1% FPR: 0.59% Acc: 99.81% | 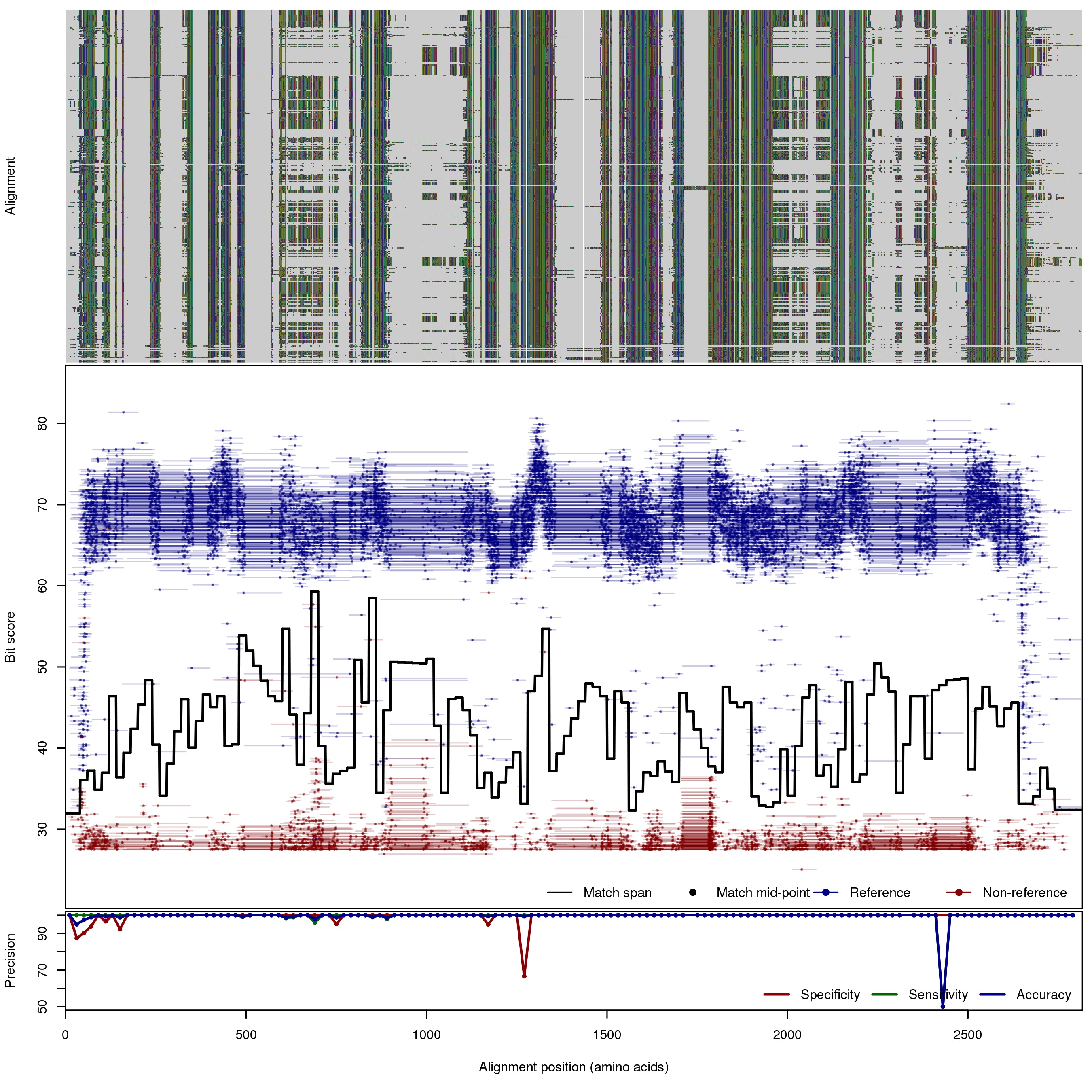 | Download Search online | |||
| 150bp | FNR: 0.13% FPR: 0.01% Acc: 99.98% | 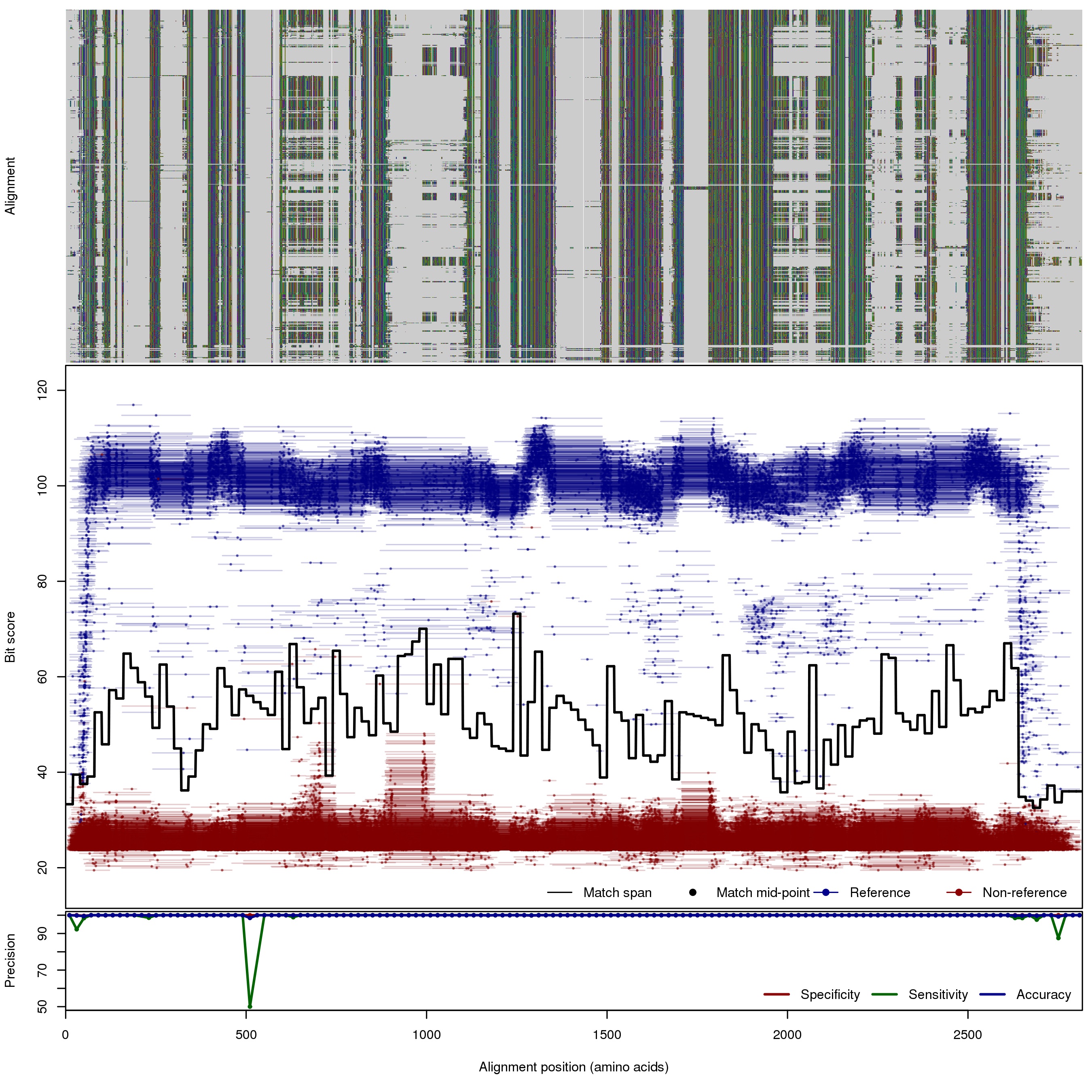 | Download Search online | |||
| 200bp | FNR: 0.22% FPR: 0.01% Acc: 99.98% | 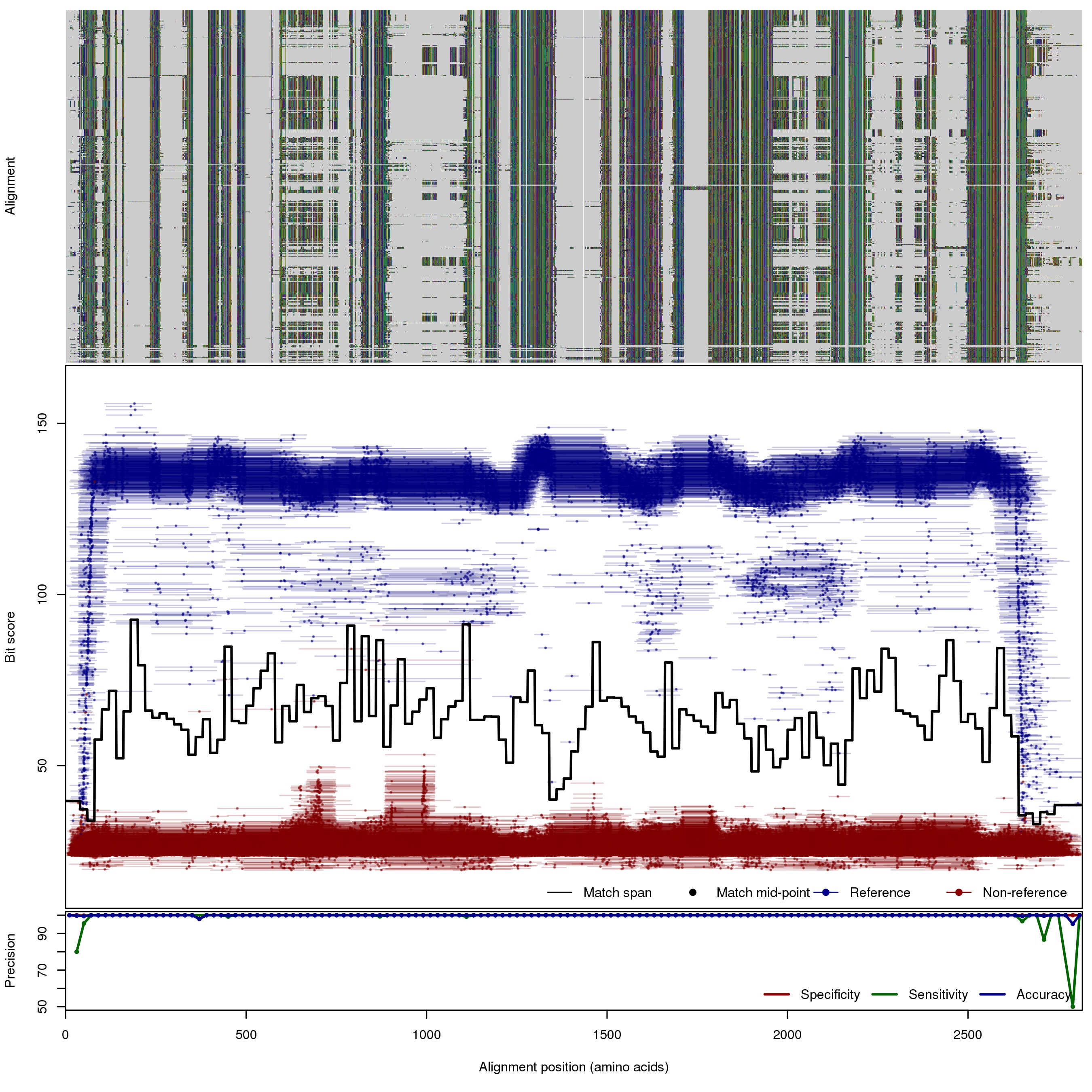 | Download Search online | |||
| 250bp | FNR: 0.17% FPR: 0% Acc: 99.99% | 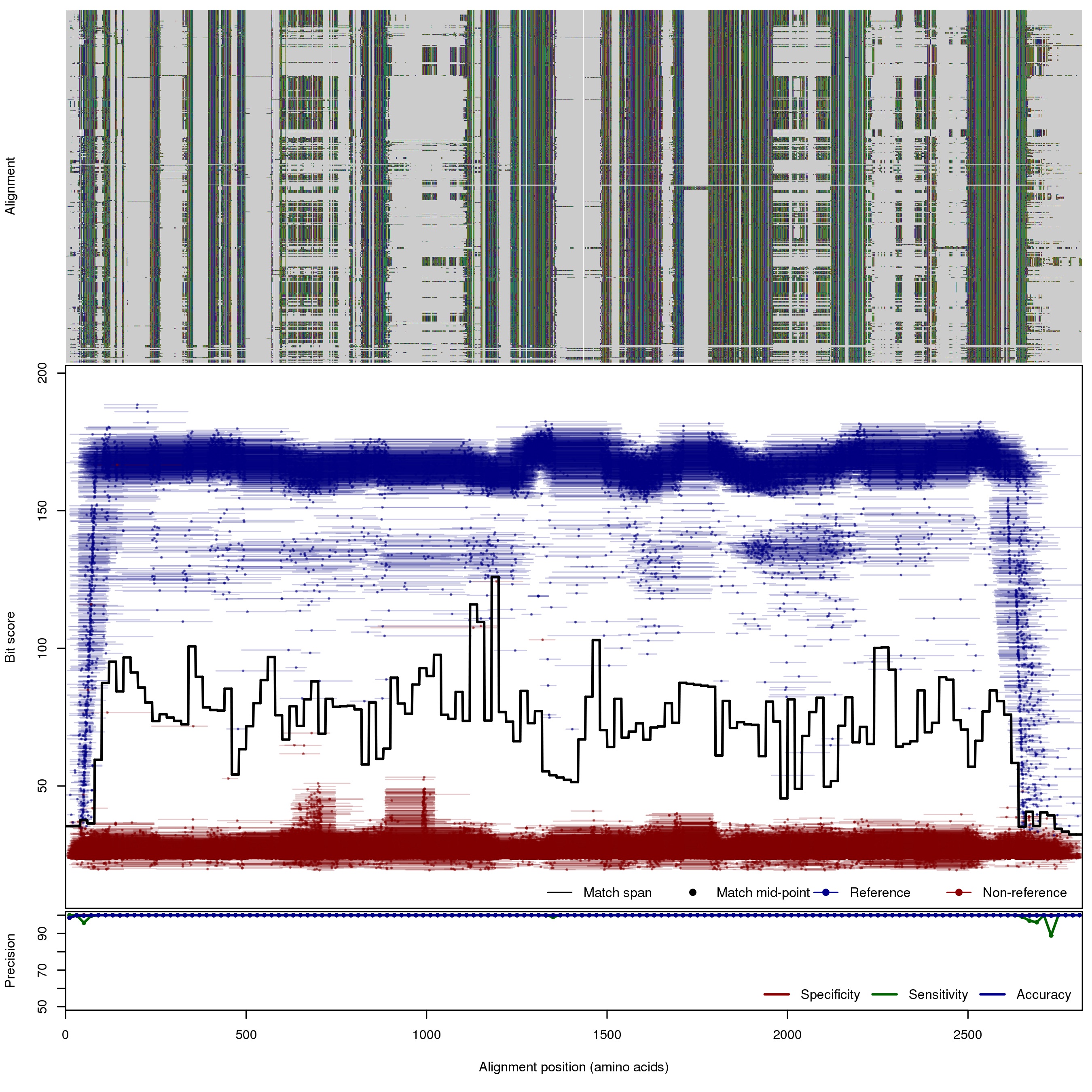 | Download Search online | |||
| 500bp | FNR: 0.29% FPR: 0% Acc: 99.99% | 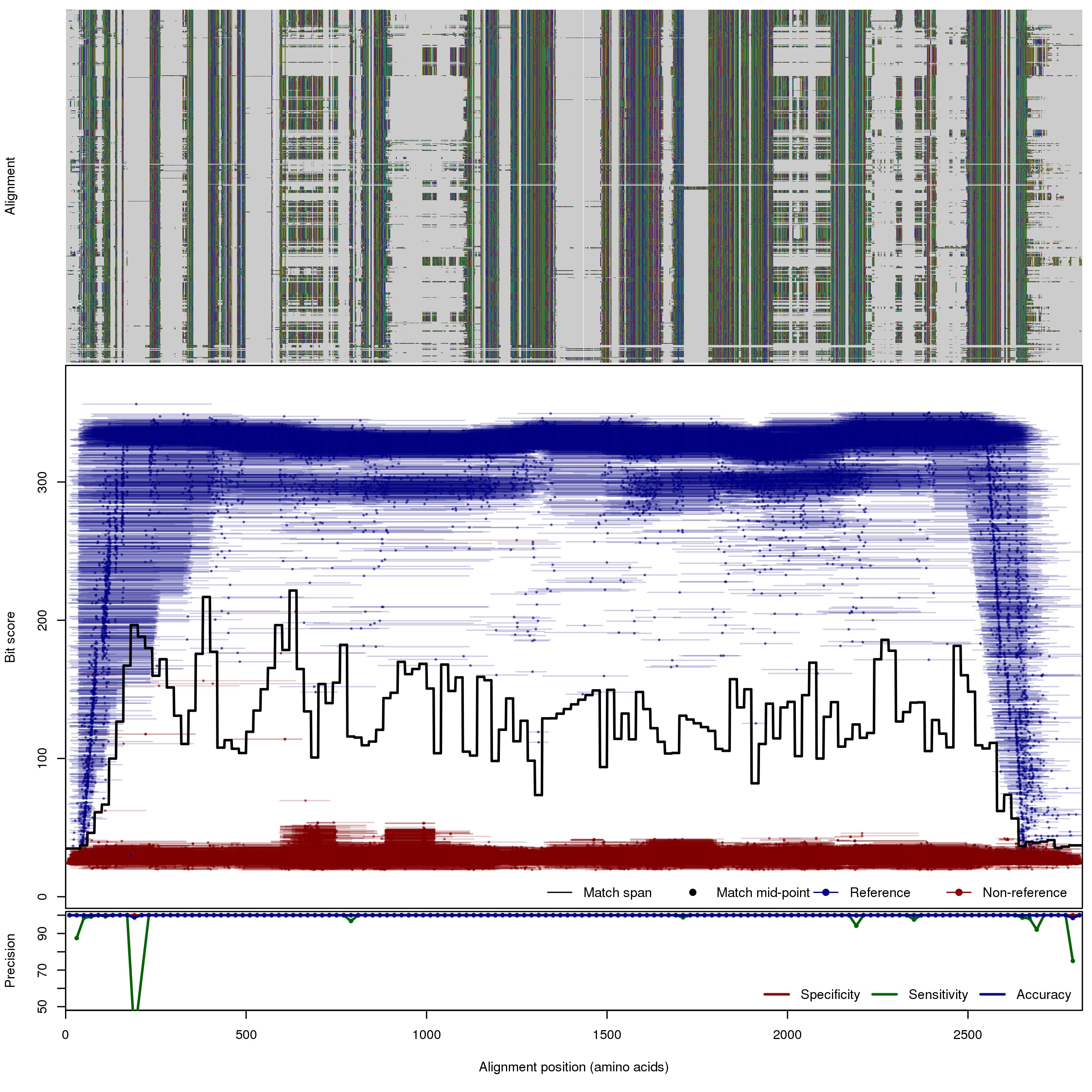 | Download Search online | |||
| TetM: Tetracycline resistance | ||||||
| TetM | 100bp | - | 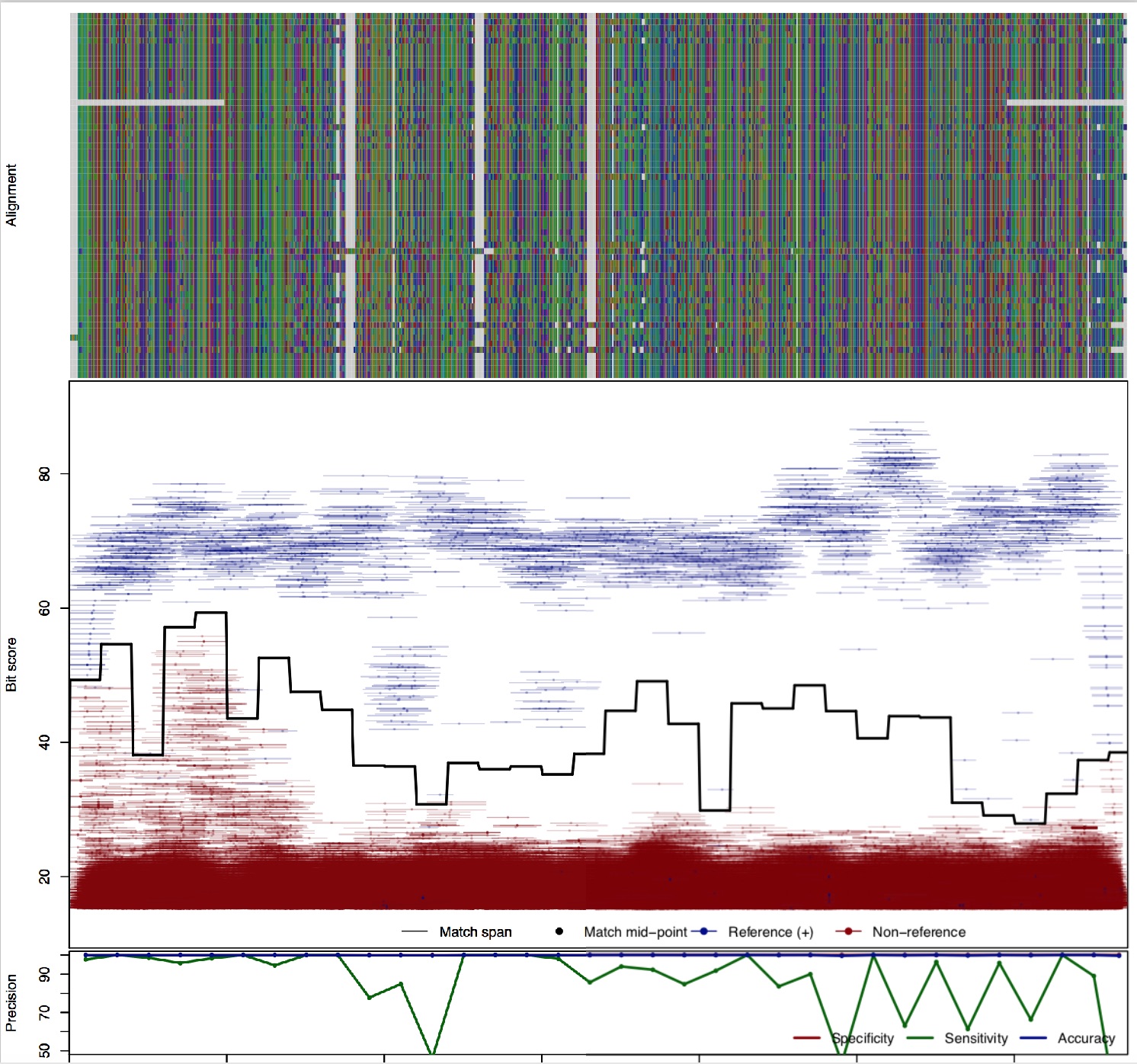 | Download Search online | Positive: 60 proteins Negative: 4 proteins Reference sequences | Last update: Apr-10-2018 |
| 150bp | - | 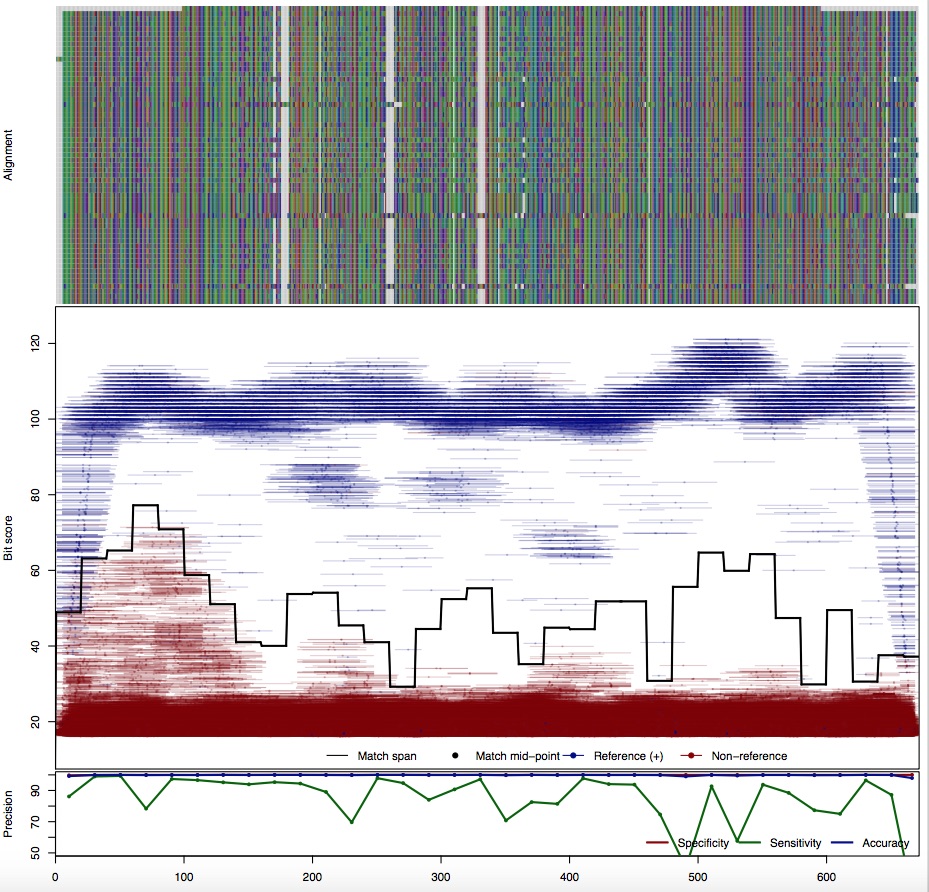 | Download Search online | |||
| UreC: Urease subunit alpha | ||||||
| UreC | 125bp | FNR: 0.58% FPR: 0.01% Acc: 99.98% | 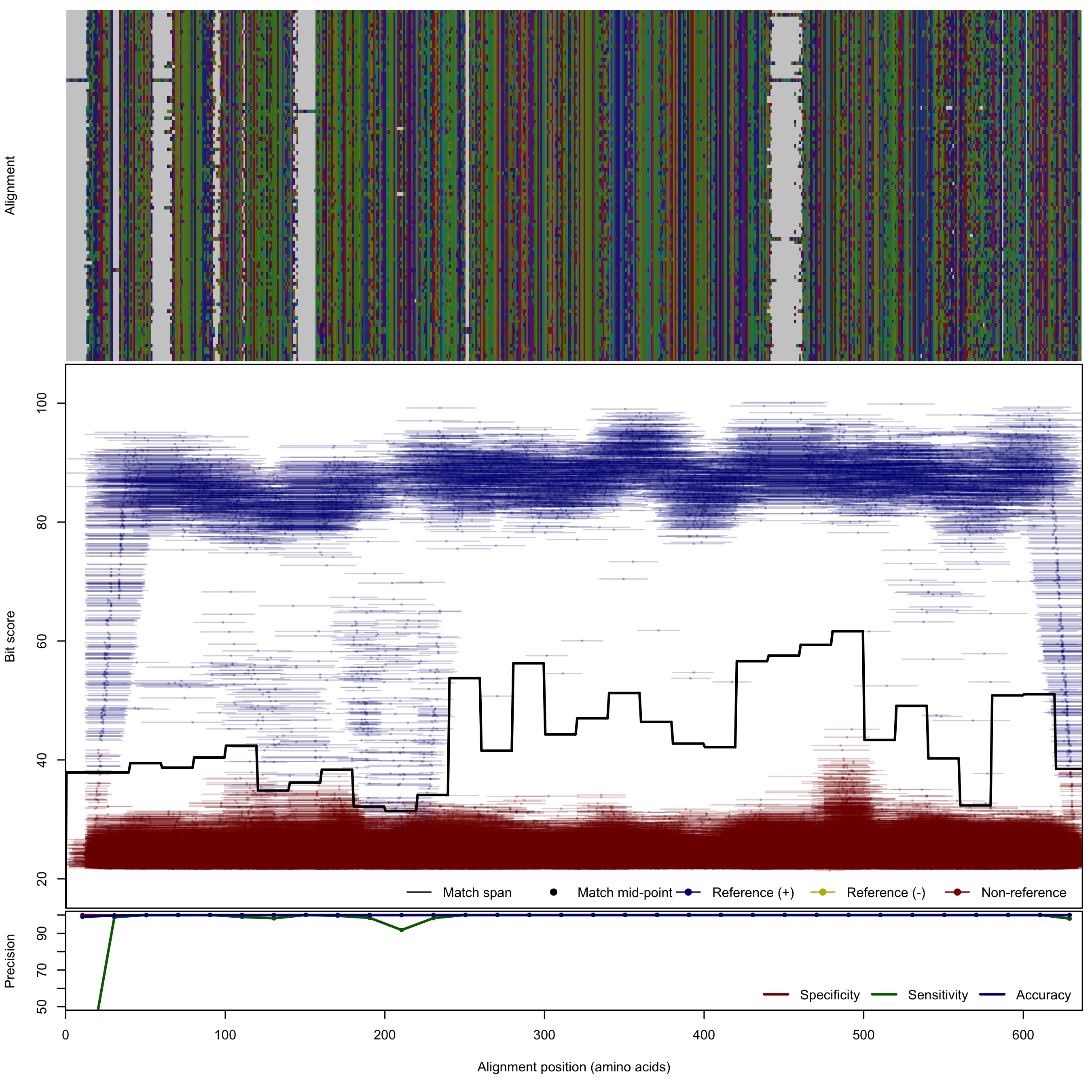 | Download Search online | Positive: 103 proteins Reference sequences | Last update: Apr-18-2017 |